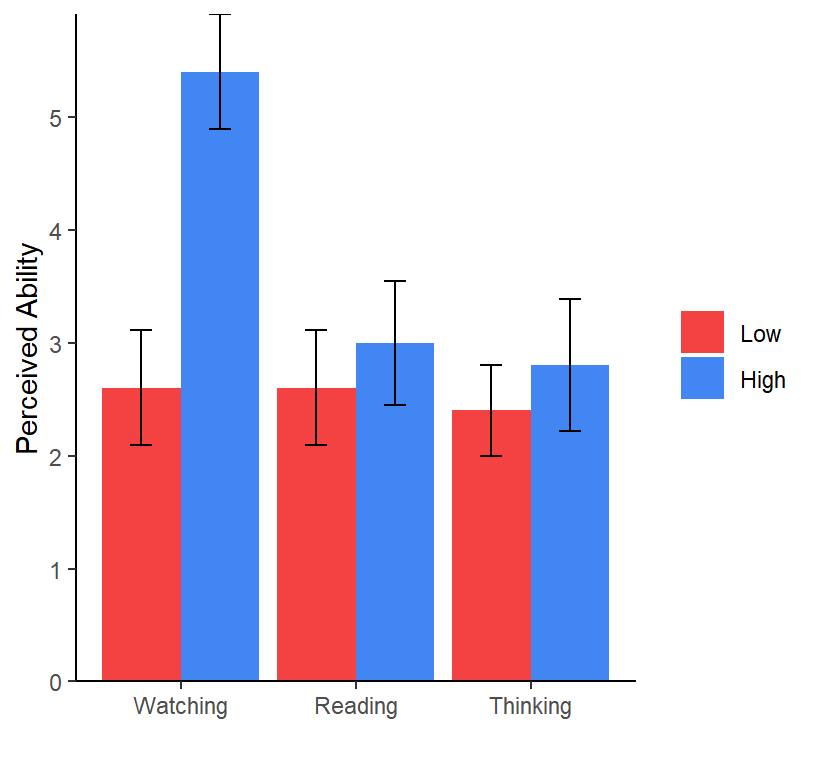
They did not hypothesize following up the other way, but we will do
it anyway as an example of how to do it. The problem we have is that we
have three level: Watching vs Reading vs Thinking @ Low, Watching vs
Reading vs Thinking @ High. In the old school approach, we first have to
run an F-test (one-way ANOVA) at each level of Level of Exposure and run
pairwise or contrasts to follow up if the One-way was significant.
Fit Model via emmeans
First, we must cut the ANOVA we calculated the way
we want to test the results:
One_way.Level<-joint_tests(Anova.Results, by = "Level")
One_way.Level
## Level = Low:
## model term df1 df2 F.ratio p.value
## Type 2 24 0.051 0.9507
##
## Level = High:
## model term df1 df2 F.ratio p.value
## Type 2 24 7.949 0.0022
Because there are three level you cannot ever run these as t-tests.
Also, the package will not easily give you effect sizes. Luckly with
between-subject ANOVAs its easy to convert your F values over to effect
sizes. Note: This will NOT work the same was a more complex designs
(mixed or repeated measures).
You can convert the F values into \(\eta_{p}^2\). [Formula from your
textbook]
\[\eta_{p}^2 =
\frac{df_bF}{df_bF+df_w}\] Note: Since we have 2 F values, we can
get R to give us 2 effect sizes!
Fs = One_way.Level$F.ratio
df_b = One_way.Level$df1
df_w =One_way.Level$df2
eta_p_exposure<- (Fs * df_b)/(Fs * df_b + df_w)
eta_p_exposure
## [1] 0.004232014 0.398466089
Simple effect for Low, F(2,24) = 0.051, p <
0.9507, \(\eta^2_p\) = 0.0042
Simple effect for High, F(2,24) = 7.949, **p* < 0.0022,
\(\eta^2_p\) = 0.3985
So we have a simple effect of High exposure. We would need to follow
this effect up to see which is different. The degree of correction you
use will depend on if its planned or unplanned comparison.
Follow up of Significant Simple Effect
The code will run these are 2 families of 3 tests. You need to ignore
the results of the low exposure group.
Specific Contrasts
Pairwise approach, specific to only significant simple effect you
want to follow up (and apply some correction)
Simple.Effects.at.Level<-emmeans(Anova.Results, ~Type+Level)
Simple.Effects.at.Level
Set2 <- list(
High.EvsR = c(0,0,0,-1,1,0),
High.WvsT = c(0,0,0,-1,0,1),
High.RvsT = c(0,0,0,0,-1,1))
contrast(Simple.Effects.at.Level,Set2,adjust='MVT')
## Type Level emmean SE df lower.CL upper.CL
## Watching Low 2.6 0.513 24 1.54 3.66
## Reading Low 2.6 0.513 24 1.54 3.66
## Thinking Low 2.4 0.513 24 1.34 3.46
## Watching High 5.4 0.513 24 4.34 6.46
## Reading High 3.0 0.513 24 1.94 4.06
## Thinking High 2.8 0.513 24 1.74 3.86
##
## Confidence level used: 0.95
## contrast estimate SE df t.ratio p.value
## High.EvsR -2.4 0.726 24 -3.307 0.0081
## High.WvsT -2.6 0.726 24 -3.583 0.0042
## High.RvsT -0.2 0.726 24 -0.276 0.9591
##
## P value adjustment: mvt method for 3 tests
Simplier, but less control
It will correct for 3 tests per family (almost same at MVT). Note its
corrected per family. So you would need to ignore the low group.
Simple.Effects.by.Level<-emmeans(Anova.Results, ~Type|Level)
pairs(Simple.Effects.by.Level,adjust='tukey')
## Level = Low:
## contrast estimate SE df t.ratio p.value
## Watching - Reading 0.0 0.726 24 0.000 1.0000
## Watching - Thinking 0.2 0.726 24 0.276 0.9591
## Reading - Thinking 0.2 0.726 24 0.276 0.9591
##
## Level = High:
## contrast estimate SE df t.ratio p.value
## Watching - Reading 2.4 0.726 24 3.307 0.0080
## Watching - Thinking 2.6 0.726 24 3.583 0.0041
## Reading - Thinking 0.2 0.726 24 0.276 0.9591
##
## P value adjustment: tukey method for comparing a family of 3 estimates
“Complex” Linear Contrast
Here you can merge and compare groups as before. You can apply
corrections if you lots of tests, but they will be within the family:
Bonferroni, Sidak, mvt, or FDR.
Simple.Effects.at.Level<-emmeans(Anova.Results, ~Type+Level)
Set3 <- list(
WvsRT = c(0,0,0,1,-.5,-.5))
contrast(Simple.Effects.at.Level,Set3,adjust='none')
## contrast estimate SE df t.ratio p.value
## WvsRT 2.5 0.628 24 3.978 0.0006
“Consecutive” Contrast
If the data were ordinal, you could test them in some kind of logical
order. You can apply corrections if you lots of tests, but they will be
within the family: Bonferroni, Sidak, mvt, or FDR.
Consec.Set <- list(
WvsR = c(-1,1,0),
RvsT = c(0,-1,1))
contrast(Simple.Effects.by.Level,Consec.Set,adjust='none')
# Also
# contrast(Simple.Effects.By.Level,'consec',adjust='none')
## Level = Low:
## contrast estimate SE df t.ratio p.value
## WvsR 0.0 0.726 24 0.000 1.0000
## RvsT -0.2 0.726 24 -0.276 0.7852
##
## Level = High:
## contrast estimate SE df t.ratio p.value
## WvsR -2.4 0.726 24 -3.307 0.0030
## RvsT -0.2 0.726 24 -0.276 0.7852
“Polynomial” Contrast
If the data were ordinal, you could test to see if they have a linear
slope or curvilinear slope. You can apply corrections if you lots of
tests, but they will be within the family: Bonferroni, Sidak, mvt, or
FDR.
Poly.Set <- list(
Linear = c(-1,0,1),
Quad = c(-.5,1,-.5))
contrast(Simple.Effects.by.Level,Poly.Set,adjust='none')
# Also
# contrast(Simple.Effects.By.Level,'poly',adjust='none')
## Level = Low:
## contrast estimate SE df t.ratio p.value
## Linear -0.2 0.726 24 -0.276 0.7852
## Quad 0.1 0.628 24 0.159 0.8749
##
## Level = High:
## contrast estimate SE df t.ratio p.value
## Linear -2.6 0.726 24 -3.583 0.0015
## Quad -1.1 0.628 24 -1.750 0.0929