Non-Linear Fit Types that can be transformed into linear regression
(basic types)
- Polynomials: Power & Orthogonal
- Growth: Exponential, Power, Logarithmic
- Rates: Reciprocal
- Correlations
Power Polynomials
- Models that simply curves, such as quadratic or cubic effects
- Linear: \(Y =B_{1}X + B_0\)
- Quadratic: \(Y = B_{1.2}X + B_{2.1}X^2 +
B_0\)
- Cubic: \(Y = B_{1.23}X + B_{2.13}X^2 +
B_{3.12}X^3 + B_0\)
Simulate a Exam Score ~ Anxiety
\(Y = 17.5X -2.5X^2 + 40 +
\epsilon\)
Where, \(X\) = Uniform distribution
of Likert scores between 0 to 7 (0 = No anxiety) & \(\epsilon\) = Random Normal [\(M = 0\) & \(S=10\)]
set.seed(42)
n <- 250 # 250 people
x <- runif(n, 0, 7)
# Our equation to create Y
y <- -2.5*x^2 + 17.5*x+40+rnorm(n, sd=10)
#Build our data frame
Quad.Data<-data.frame(Anxiety=x,ExamScore=y)
Visualize Simulation/Data
Beyond visualizing the scatter plot, we need some way to estimate if
the effect might be linear or non-linear
Loess Line
LOESS = Locally Weighted
Scatterplot Smoothing
Does lots of regressions on small sections of the scatter plot to get
the best fit (curvy) line. Allows you visualize complex relationships,
but it can overfit the relationship (so be careful. Thus its a
good diagnostic tool, but rarely used in papers.
library(ggpubr) #graph data
ggscatter(Quad.Data, x = "Anxiety", y = "ExamScore",
add = "loess", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = FALSE, # Add correlation coefficient. see ?stat_cor
size = 1 # Size of dots
)
Fitting Power Polynomials
Forward Selection Approach
- Model \(1\): Linear fit
- Model \(2\): Quadratic fit
- Check the change in \(R^2\) and
select the best fit models
- Model \(i\): keep going up until
you are satisfied
- Note: Higher order terms ALWAYS increase \(R^2\), you need to make sure it is both a
significant improvement and meaningful improvement
- in R there are two ways to add Power Polynomials: I(Term^2)
or using the poly command
#Linear model
Model.1<-lm(ExamScore~Anxiety, data= Quad.Data)
#Quad model
Model.2<-lm(ExamScore~Anxiety+I(Anxiety^2), data= Quad.Data)
- Test to see if the model is improved by the new term
anova(Model.1,Model.2)
## Analysis of Variance Table
##
## Model 1: ExamScore ~ Anxiety
## Model 2: ExamScore ~ Anxiety + I(Anxiety^2)
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 248 43354
## 2 247 21319 1 22036 255.31 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
|
|
|
|
Dependent variable:
|
|
|
|
|
|
ExamScore
|
|
|
Linear
|
Quadratic
|
|
|
(1)
|
(2)
|
|
|
|
Constant
|
60.333*** (1.686)
|
39.681*** (1.753)
|
|
Anxiety
|
-0.137 (0.411)
|
17.753*** (1.156)
|
|
I(Anxiety2)
|
|
-2.559*** (0.160)
|
|
|
|
Observations
|
250
|
250
|
|
R2
|
0.0004
|
0.508
|
|
Adjusted R2
|
-0.004
|
0.505
|
|
Residual Std. Error
|
13.222 (df = 248)
|
9.290 (df = 247)
|
|
F Statistic
|
0.111 (df = 1; 248)
|
127.767*** (df = 2; 247)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
- The change in \(R^2\) = 0.5080438
and we can see that the change was significant
- Would the third order term be any better?
#Cubic model
Model.3<-lm(ExamScore~Anxiety+I(Anxiety^2)+I(Anxiety^3), data= Quad.Data)
anova(Model.1,Model.2,Model.3)
## Analysis of Variance Table
##
## Model 1: ExamScore ~ Anxiety
## Model 2: ExamScore ~ Anxiety + I(Anxiety^2)
## Model 3: ExamScore ~ Anxiety + I(Anxiety^2) + I(Anxiety^3)
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 248 43354
## 2 247 21319 1 22035.8 255.0382 <2e-16 ***
## 3 246 21255 1 63.8 0.7378 0.3912
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
|
|
|
|
Dependent variable:
|
|
|
|
|
|
ExamScore
|
|
|
Linear
|
Quadratic
|
Cubic
|
|
|
(1)
|
(2)
|
(3)
|
|
|
|
Constant
|
60.333*** (1.686)
|
39.681*** (1.753)
|
38.386*** (2.312)
|
|
Anxiety
|
-0.137 (0.411)
|
17.753*** (1.156)
|
20.023*** (2.885)
|
|
I(Anxiety2)
|
|
-2.559*** (0.160)
|
-3.371*** (0.959)
|
|
I(Anxiety3)
|
|
|
0.077 (0.090)
|
|
|
|
Observations
|
250
|
250
|
250
|
|
R2
|
0.0004
|
0.508
|
0.510
|
|
Adjusted R2
|
-0.004
|
0.505
|
0.504
|
|
Residual Std. Error
|
13.222 (df = 248)
|
9.290 (df = 247)
|
9.295 (df = 246)
|
|
F Statistic
|
0.111 (df = 1; 248)
|
127.767*** (df = 2; 247)
|
85.333*** (df = 3; 246)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
Second order power polynomial was our best fit
Model 2 in Detail
Since we simulated our model we know what the answers
should be \(Y = 17.5X -2.5X^2
+ 40 +\epsilon\)
- Extract our terms from our regression and compare to our simulated
values.
IT<-round(Model.2$coefficients[1],2)
LT<-round(Model.2$coefficients[2],2)
QT<-round(Model.2$coefficients[3],2)
Regression estimated values: \(Y =\)
17.75\(X\) -2.56\(X^2 +\) 39.68 \(+\epsilon\)
- Our regression did a good job, but what if our sample was
smaller?
- Redo our analysis with n = 5 to 60 and see how the terms change [see
simulation in separate R file]
- the gray band = 95% CI
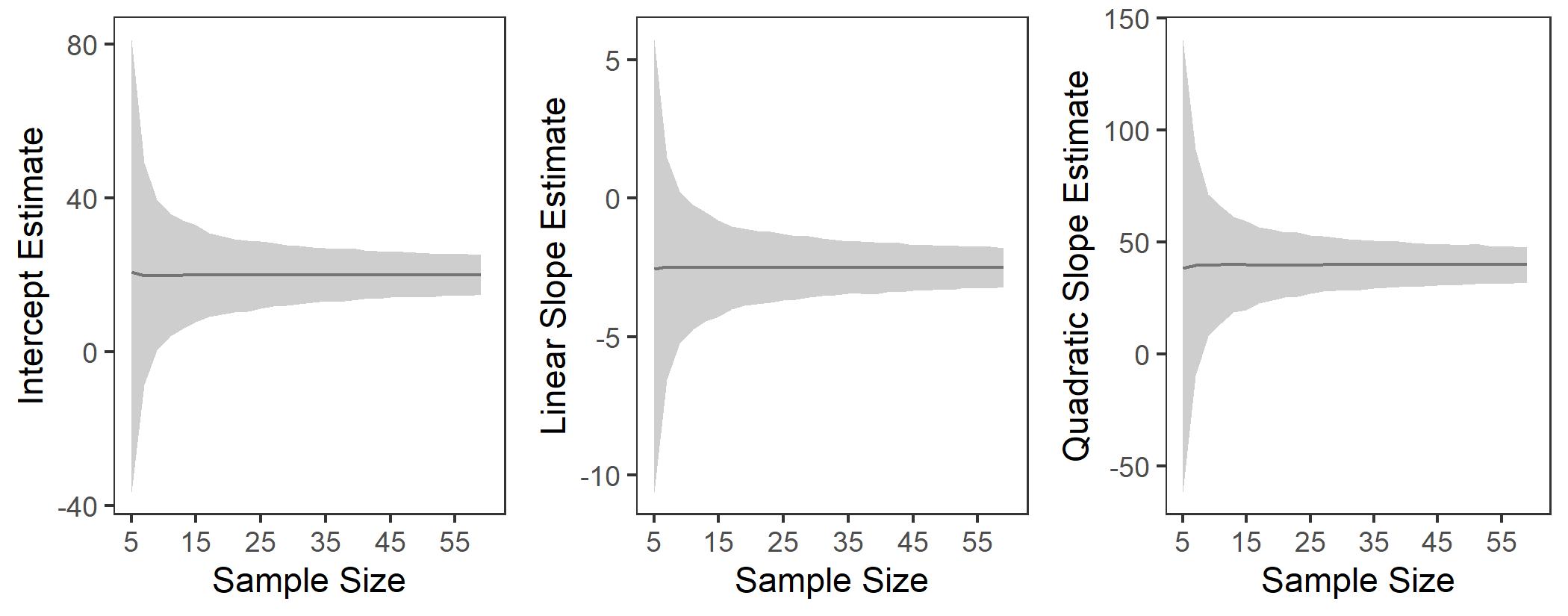
It’s important to know that in small samples polynomial estimates can
be very unstable. Also note: If you flip the quadratic term, the
line will flip direction!
How to plot this function (and not the loess line)
- You can solve the regression equation for different values of x
- in R you can call the
effects package to do work for
you to solve the regression equation at different values of \(X\) [0 to 7].
library(effects)
plot(effect("Anxiety", Model.2,
xlevels=list(Anxiety=seq(0,7,1))))
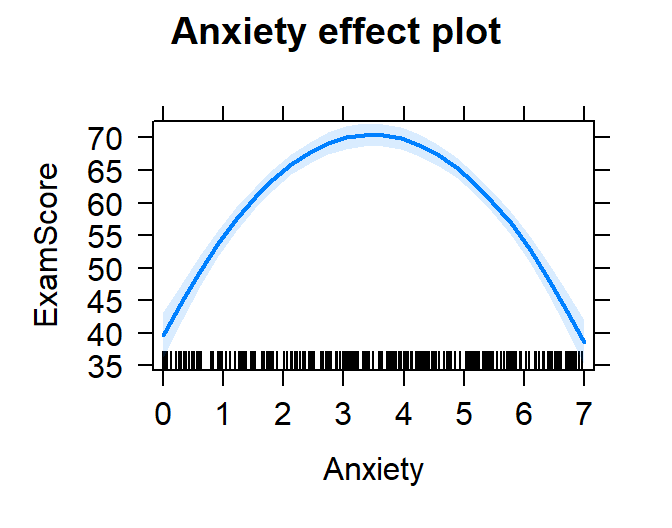
Code approach to power polynomials
You can use a command called poly which will do both
power and orthogonal polynomials. But you cannot have missing data to
use this code. The function has serval important arguments we will focus
on: poly(x, degree = 1, raw = FALSE, simple=FALSE). degree
= order number. So degree = 3 tests the linear, quadratic,
and cubic. raw = FALSE tests the orthogonal versions. To
keep them as power polynomials we must say raw = TRUE.
Lastly, there is simple=FALSE, this is the default and
keeps the attributes assigned the new vectors you have created. I
suggest leaving this alone, meaning leave the default as is, but it is
useful to know as sometimes when we pass the results of our model this
can create problems with other packages (but I have yet to see
this).
#Cubic model
Model.3.P<-lm(ExamScore~poly(Anxiety,3,raw=TRUE), data= Quad.Data)
summary(Model.3.P)
##
## Call:
## lm(formula = ExamScore ~ poly(Anxiety, 3, raw = TRUE), data = Quad.Data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -25.7936 -6.7479 -0.0587 6.5252 24.9911
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 38.38640 2.31249 16.600 < 2e-16 ***
## poly(Anxiety, 3, raw = TRUE)1 20.02317 2.88488 6.941 3.44e-11 ***
## poly(Anxiety, 3, raw = TRUE)2 -3.37082 0.95900 -3.515 0.000523 ***
## poly(Anxiety, 3, raw = TRUE)3 0.07736 0.09006 0.859 0.391190
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 9.295 on 246 degrees of freedom
## Multiple R-squared: 0.51, Adjusted R-squared: 0.504
## F-statistic: 85.33 on 3 and 246 DF, p-value: < 2.2e-16
Above you can see the names of the paramaters in the model have
changed.
- poly(Anxiety, 3, raw = TRUE)1 = linear
- poly(Anxiety, 3, raw = TRUE)2 = quadratic
- poly(Anxiety, 3, raw = TRUE)3 = cubic.
Orthogonal Polynomials
Problem with Power polynomials is interpreting them independently.
The higher order terms depend on, the lower order terms. Thus linear
term can be significant cause the higher order term is significant, but
there is no linear trend in the data (just like in today’s example).
This is because power polynomials correlate with each other
(they are are not unique predictors)
Linear.Values<-c(1,2,3,4,5,6,7)
Quad.Values<-Linear.Values^2
L.vs.Q<-data.frame(Linear.Values=Linear.Values,Quad.Values=Quad.Values)
ggscatter(L.vs.Q, x = "Linear.Values", y = "Quad.Values",
add = "reg.line", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = TRUE, # Add correlation coefficient. see ?stat_cor
size = 2 # Size of dots
)
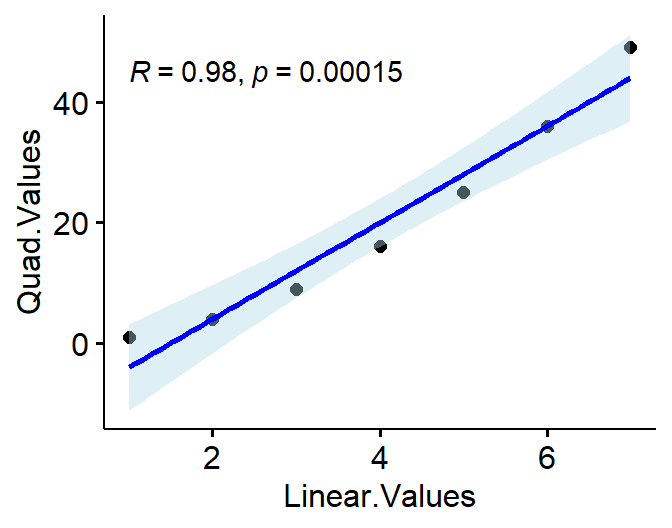
- Thus they are basically multicollinear
- Let’s run our VIF to verify
library(car)
vif(Model.2)
## Anxiety I(Anxiety^2)
## 16.05275 16.05275
Technically, it’s OK that they are multicollinear, but you cannot
interpret each term without looking at the other (the p-values will be
problematic).
- The solution to make the linear and quadratic terms correlate with
each at zero (just as we did with ANOVA)
- We can use the poly code in R to make them Orthogonal.
- Here is an example of what R is going to do:
LinearTerms<-c(1,2,3,4,5,6,7)
#make them Orthogonal
O.Poly<-poly(LinearTerms,2)
L.vs.Q.Ortho<-data.frame(Linear.Values.Ortho=O.Poly[,1],Quad.Values.Ortho=O.Poly[,2])
ggscatter(L.vs.Q.Ortho, x = "Linear.Values.Ortho", y = "Quad.Values.Ortho",
add = "reg.line", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = TRUE, # Add correlation coefficient. see ?stat_cor
size = 2 # Size of dots
)
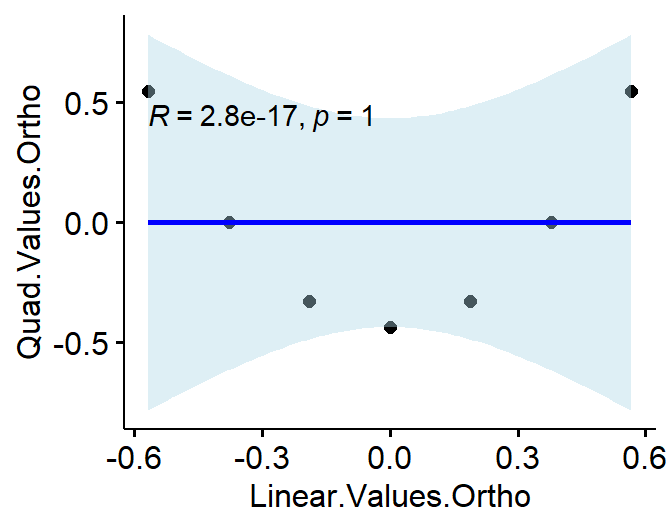
Correlation between linear and quadratic is now, r = 0!
#lets make a new poly vector
Model.1.O<-lm(ExamScore~poly(Anxiety, 1), data= Quad.Data)
Model.2.O<-lm(ExamScore~poly(Anxiety, 2), data= Quad.Data)
anova(Model.1.O,Model.2.O)
## Analysis of Variance Table
##
## Model 1: ExamScore ~ poly(Anxiety, 1)
## Model 2: ExamScore ~ poly(Anxiety, 2)
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 248 43354
## 2 247 21319 1 22036 255.31 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
|
|
|
|
Dependent variable:
|
|
|
|
|
|
ExamScore
|
|
|
Linear.Ortho
|
Quadratic.Ortho
|
|
|
(1)
|
(2)
|
|
|
|
Constant
|
59.845*** (0.836)
|
59.845*** (0.588)
|
|
poly(Anxiety, 1)
|
-4.403 (13.222)
|
|
|
poly(Anxiety, 2)1
|
|
-4.403 (9.290)
|
|
poly(Anxiety, 2)2
|
|
-148.444*** (9.290)
|
|
|
|
Observations
|
250
|
250
|
|
R2
|
0.0004
|
0.508
|
|
Adjusted R2
|
-0.004
|
0.505
|
|
Residual Std. Error
|
13.222 (df = 248)
|
9.290 (df = 247)
|
|
F Statistic
|
0.111 (df = 1; 248)
|
127.767*** (df = 2; 247)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
- Notice that the linear term is not significant, but the quadratic is
- but the actual estimate of the slopes are nonsensical
- This is because you have rescaled the x-values in a
funny way (see plot above)
- But no more multicollinearity
- Let’s run our VIF to verify
- Note: poly gets read to vif as one factor, so I will manually enter
these into the model
Quad.Data$Anxiety.O.1<-poly(Quad.Data$Anxiety,2)[,1]
Quad.Data$Anxiety.O.2<-poly(Quad.Data$Anxiety,2)[,2]
Model.2.O.2<-lm(ExamScore~Anxiety.O.1+Anxiety.O.2, data= Quad.Data)
vif(Model.2.O.2)
## Anxiety.O.1 Anxiety.O.2
## 1 1
- Also the \(R^2\) do not differ
between the power or orthogonal polynomials
- Other pros of using orthogonal polynomials will come when we start
looking at interactions
Coding shortcut (Use this for your analysis)
- You don’t need to premake the vector as I did above
- You can put the poly code right into the model
- but the labeling will change
- Also much easier to makes plots of the final model this way
Model.2.O.S<-lm(ExamScore~poly(Anxiety,2), data= Quad.Data)
|
|
|
|
Dependent variable:
|
|
|
|
|
|
ExamScore
|
|
|
Quadratic.Ortho
|
|
|
|
Constant
|
59.845*** (0.588)
|
|
poly(Anxiety, 2)1
|
-4.403 (9.290)
|
|
poly(Anxiety, 2)2
|
-148.444*** (9.290)
|
|
|
|
Observations
|
250
|
|
R2
|
0.508
|
|
Adjusted R2
|
0.505
|
|
Residual Std. Error
|
9.290 (df = 247)
|
|
F Statistic
|
127.767*** (df = 2; 247)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
The effects package will “know” you used poly command
and convert the results back for you automatically
plot(effect("poly(Anxiety,2)", Model.2.O.S,
xlevels=list(Anxiety=seq(0,7,1))))
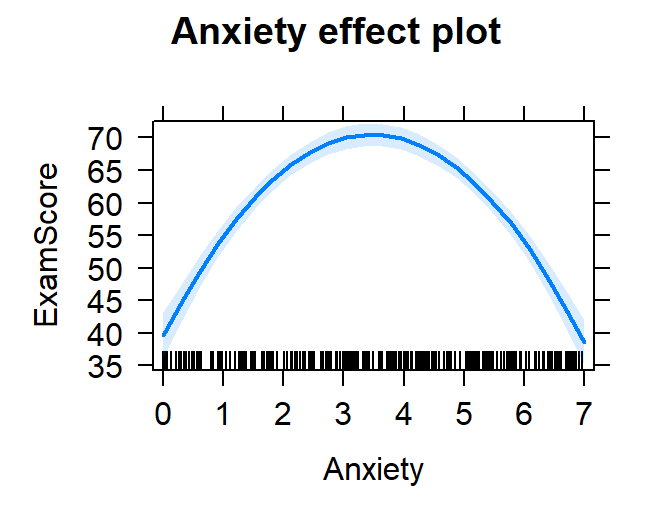
- Also, you could have said poly(Anxiety,2,raw=TRUE) to get Power
polynomial on the fly
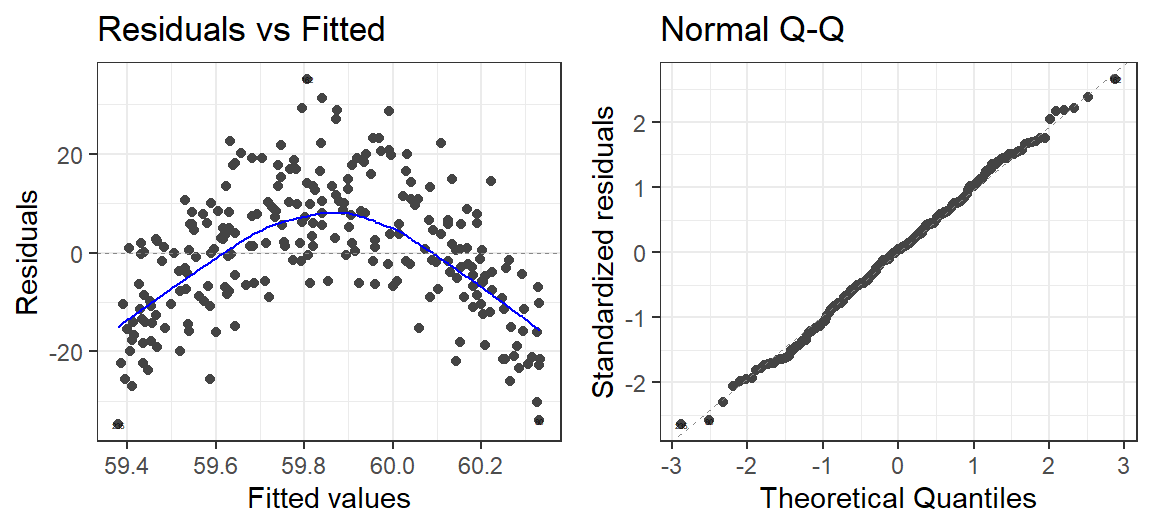
Diagnostics
- When you have the wrong number of polynomials you
will get funny looking residuals
library(ggfortify)
autoplot(Model.1, which = 1:2, label.size = 1) + theme_bw()
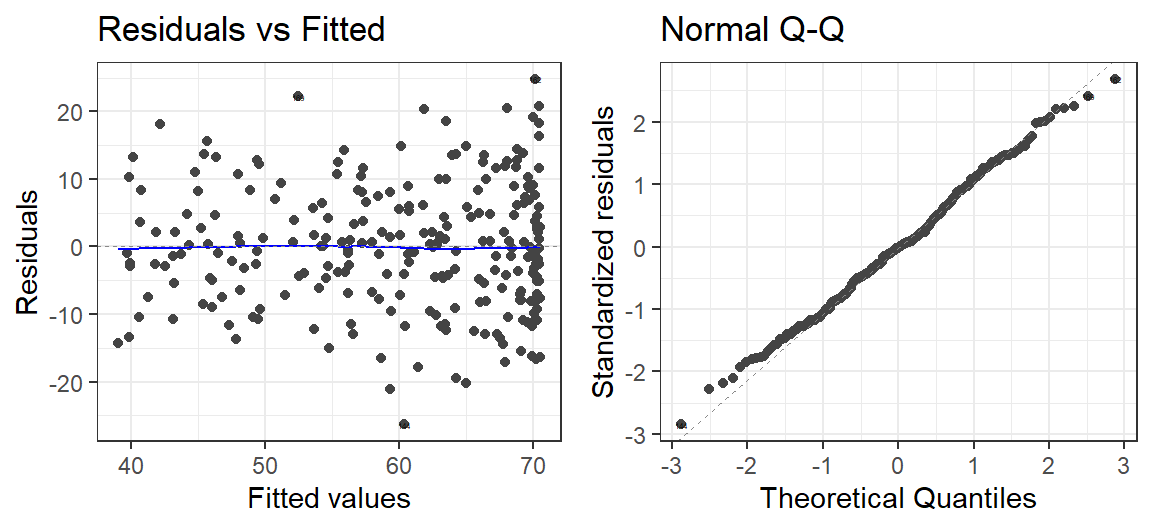
- When you have the right number of polynomials, you
will get correct looking residuals
- Here is when we use 2 when we need 2
autoplot(Model.2.O.S, which = 1:2, label.size = 1) + theme_bw()
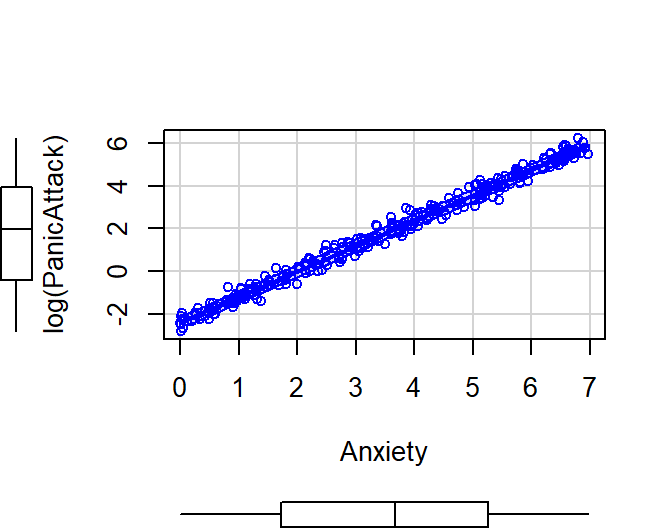
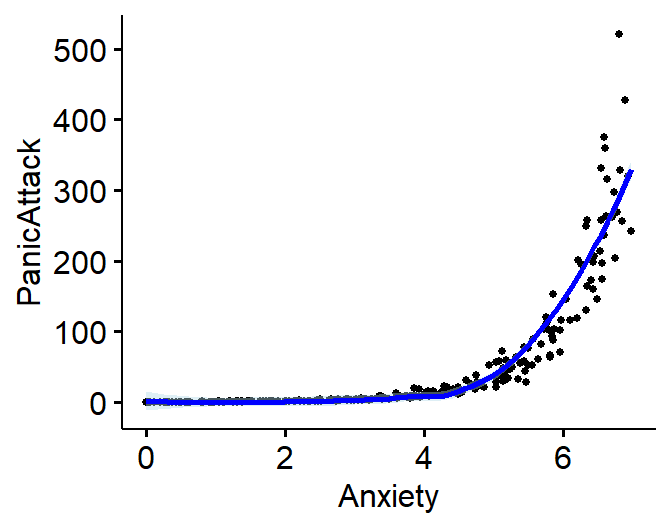
Dealing Growth Curves (Part 2)
- These are models that cannot be fit with polynomials
- Exponential growth: \(Y =
ce^{dx}\)
- Severity of panic attack (0 to 500) predicted by anxiety level
- Exponential Example: \(Y =
.09e^{1.2x+\epsilon}\)
- Note: Euler’s number, \(e =\)
2.7182818, is an irrational math constant and the base value of the
natural log
set.seed(42)
n <- 250
x <- runif(n, 0, 7)
#lets make our e value (not the error term)
e<-exp(1)
y <-.09*e^(1.20*(x+rnorm(n, sd=.25)))
Exp.Data<-data.frame(Anxiety=x,PanicAttack=y)
ggscatter(Exp.Data, x = "Anxiety", y = "PanicAttack",
add = "loess", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = FALSE, # Add correlation coefficient. see ?stat_cor
size = 1 # Size of dots
)
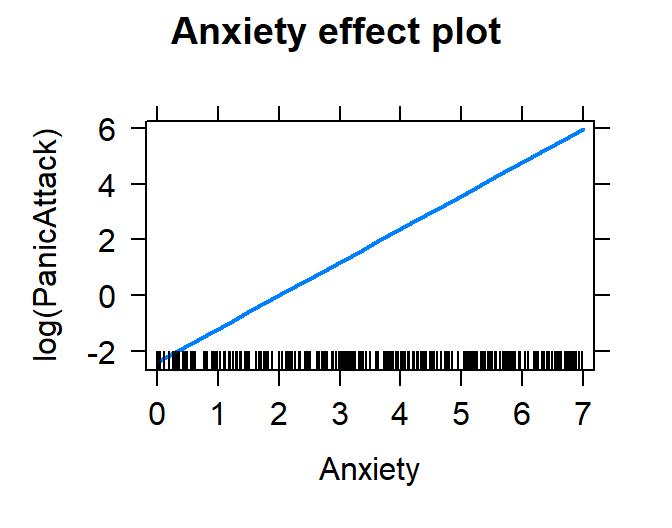
Do Polynomials fail us?
- Let’s try a second order fit since it one curve
Model.Panic.Poly<-lm(PanicAttack~poly(Anxiety,2), data= Exp.Data)
|
|
|
|
Dependent variable:
|
|
|
|
|
|
PanicAttack
|
|
|
Quadratic.Ortho
|
|
|
|
Constant
|
50.000*** (2.515)
|
|
poly(Anxiety, 2)1
|
1,006.757*** (39.762)
|
|
poly(Anxiety, 2)2
|
772.384*** (39.762)
|
|
|
|
Observations
|
250
|
|
R2
|
0.805
|
|
Adjusted R2
|
0.803
|
|
Residual Std. Error
|
39.762 (df = 247)
|
|
F Statistic
|
509.199*** (df = 2; 247)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
- Using the poly right into the package can be useful for the effects
package because we can now view independent impact of the linear and
quadratic terms
plot(effect("poly(Anxiety,2)", Model.Panic.Poly,
xlevels=list(Anxiety=seq(0,7,1))))
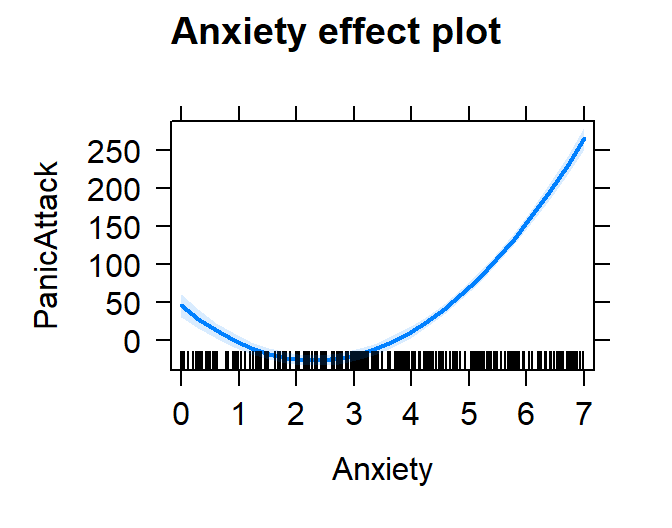 - Yikes, based on this you might tell people having zero anxiety is
worse than having lower anxiety
- Yikes, based on this you might tell people having zero anxiety is
worse than having lower anxiety
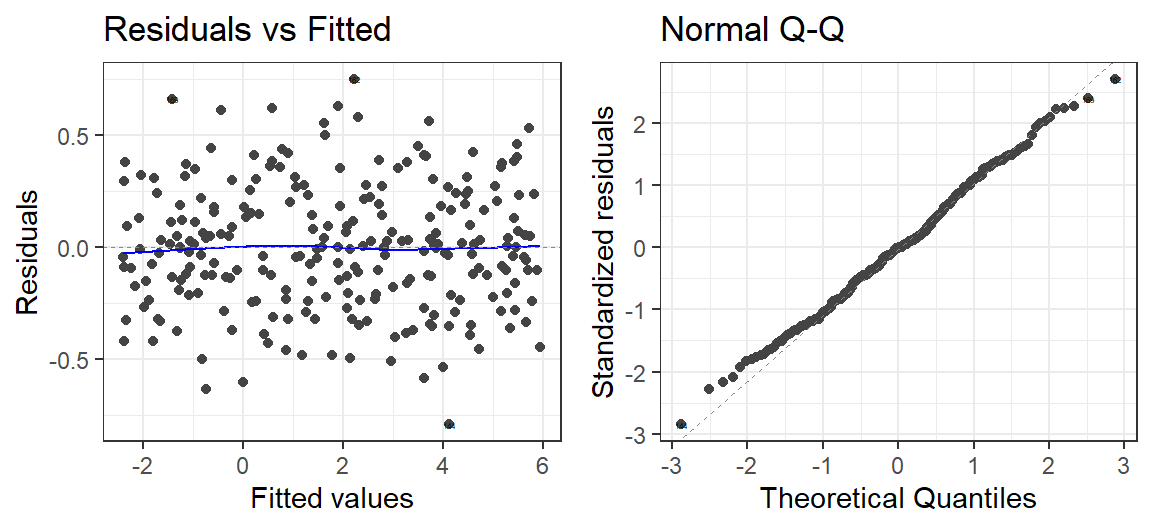
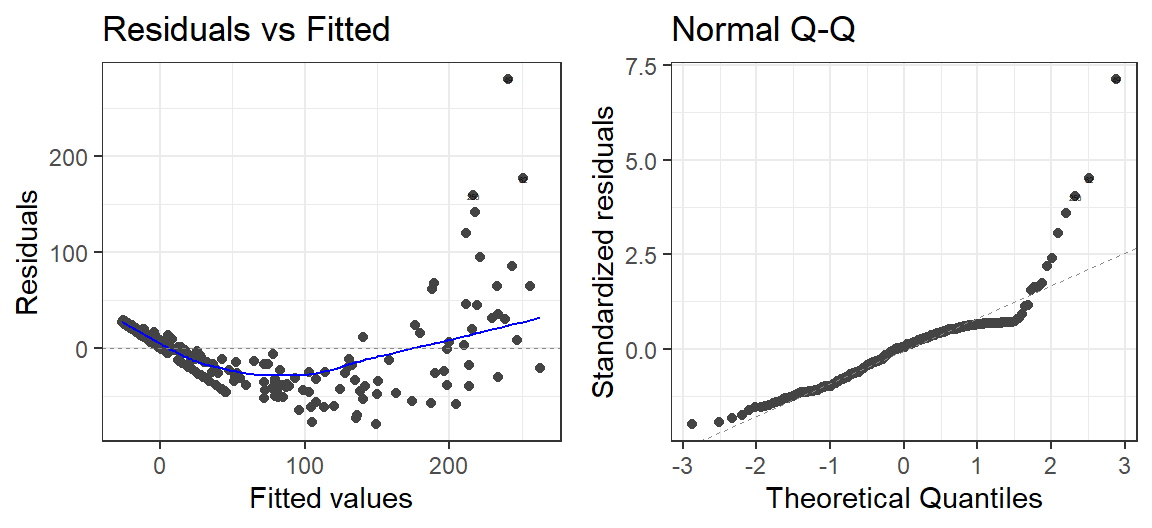
autoplot(Model.Panic.Poly, which = 1:2, label.size = 1) + theme_bw()
Power law
- Common function in motor control and perception experiments
- \(Y = cX^d\)
- Severity of sweating (0 to 100) predicted by anxiety level
- Exponential Example: \(Y =
.07(X)^{1.7}+\epsilon\)
- Note: Natural log of 0 = -inf, so if you have zeros you need to
always add 1
set.seed(42)
n <- 250
x <- runif(n, 0, 7)+1
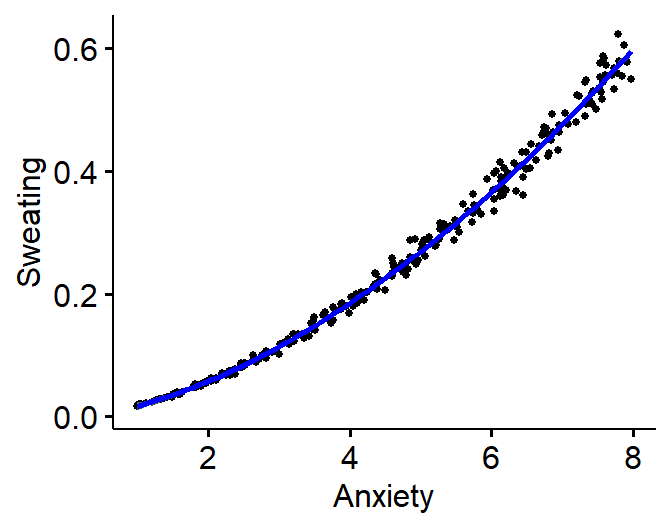
y <-.07*(x)^1.7
y = y * (.05* (5+rnorm(n, sd=.25)))
Power.Data<-data.frame(Anxiety=x,Sweating=y)
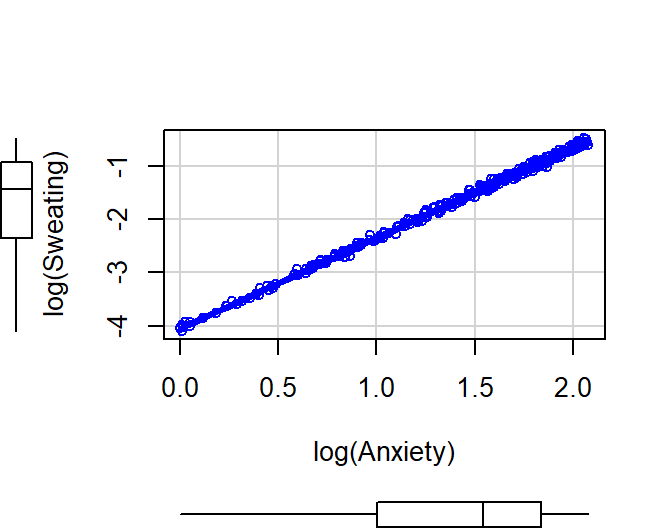
ggscatter(Power.Data, x = "Anxiety", y = "Sweating",
add = "loess", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = FALSE, # Add correlation coefficient. see ?stat_cor
size = 1 # Size of dots
)
Logarithmic
- These functions are often physical (sound intensity, earthquakes,
etc.)
- \(c^Y = dX\)
- Breathing rate (in ms between breaths) predicted by anxiety
level
- Logarithmic Example: \(Y =
.05(X*\epsilon)^{3.8}\)
set.seed(42)
n <- 250
x <- runif(n, 0, 7)+1
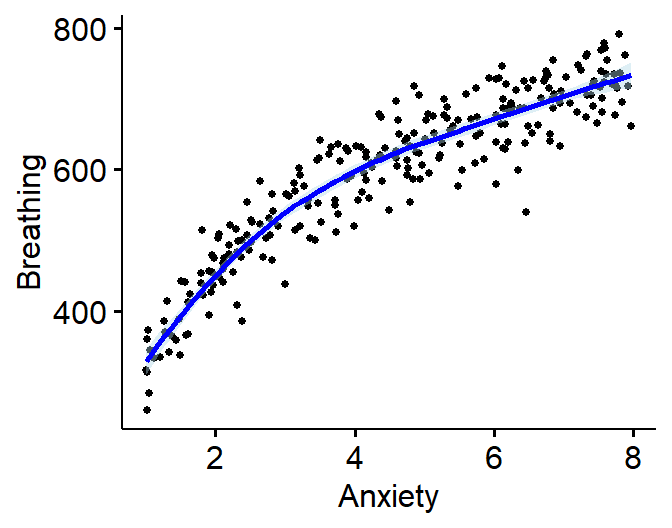
y <-200*log(x*(5+rnorm(n, sd=1)))
Logarithmic.Data<-data.frame(Anxiety=x,Breathing=y)
ggscatter(Logarithmic.Data, x = "Anxiety", y = "Breathing",
add = "loess", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = FALSE, # Add correlation coefficient. see ?stat_cor
size = 1 # Size of dots
)
- Plot Breathing by log(Anxiety)
scatterplot(Breathing~log(Anxiety), data= Logarithmic.Data)
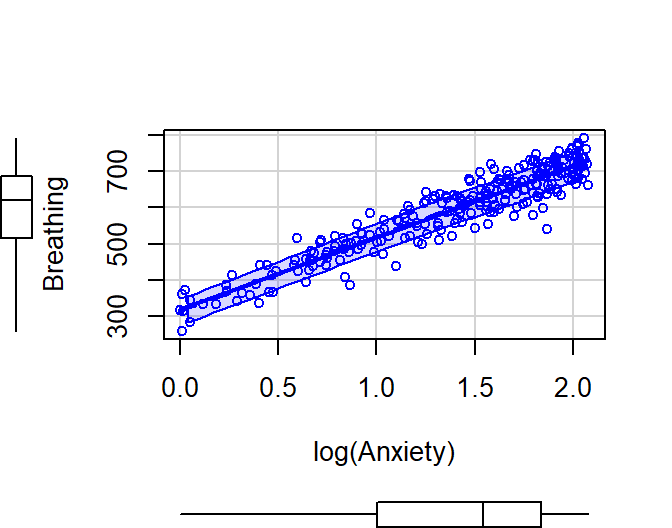
- Looks like a line now
- Let’s look at the model fit
Model.Breath.Log<-lm(Breathing~log(Anxiety), data= Logarithmic.Data)
|
|
|
|
Dependent variable:
|
|
|
|
|
|
Breathing
|
|
|
Log Transform
|
|
|
|
Constant
|
319.641*** (6.656)
|
|
log(Anxiety)
|
197.847*** (4.456)
|
|
|
|
Observations
|
250
|
|
R2
|
0.888
|
|
Adjusted R2
|
0.888
|
|
Residual Std. Error
|
38.680 (df = 248)
|
|
F Statistic
|
1,971.251*** (df = 1; 248)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
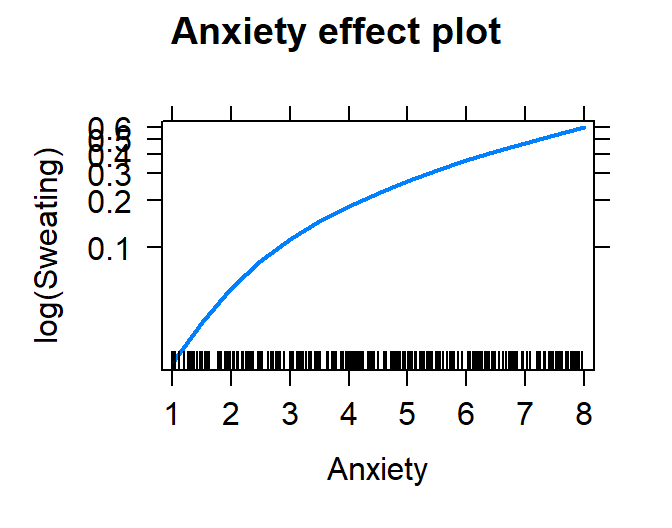
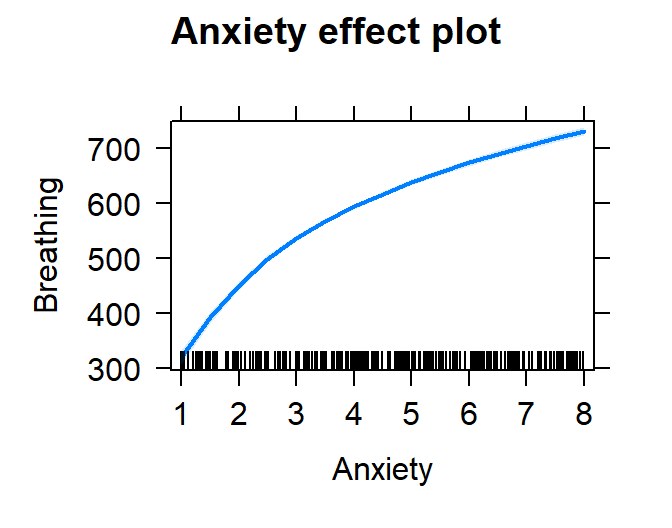
plot(effect("Anxiety", Model.Breath.Log))
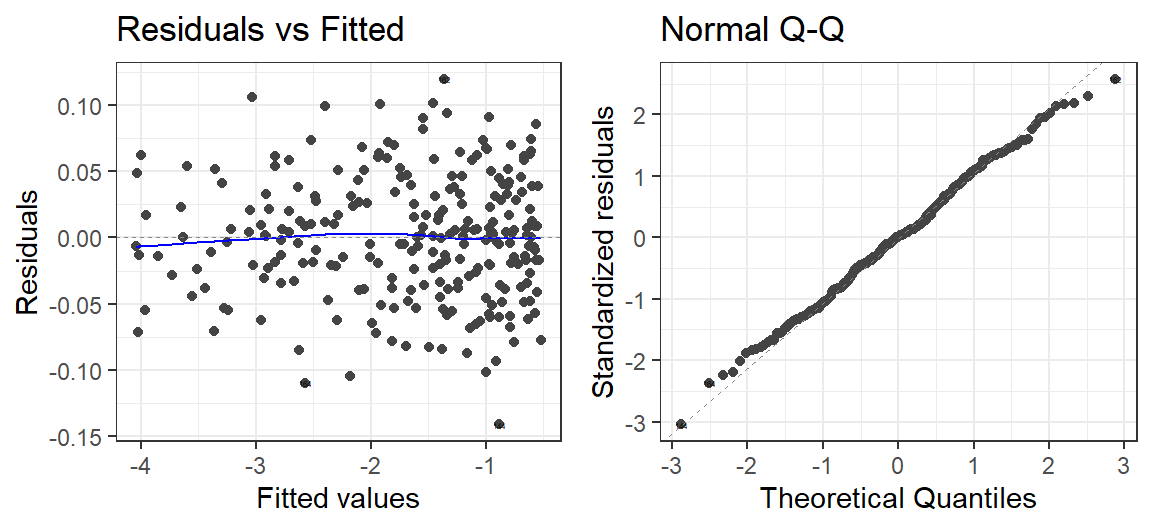
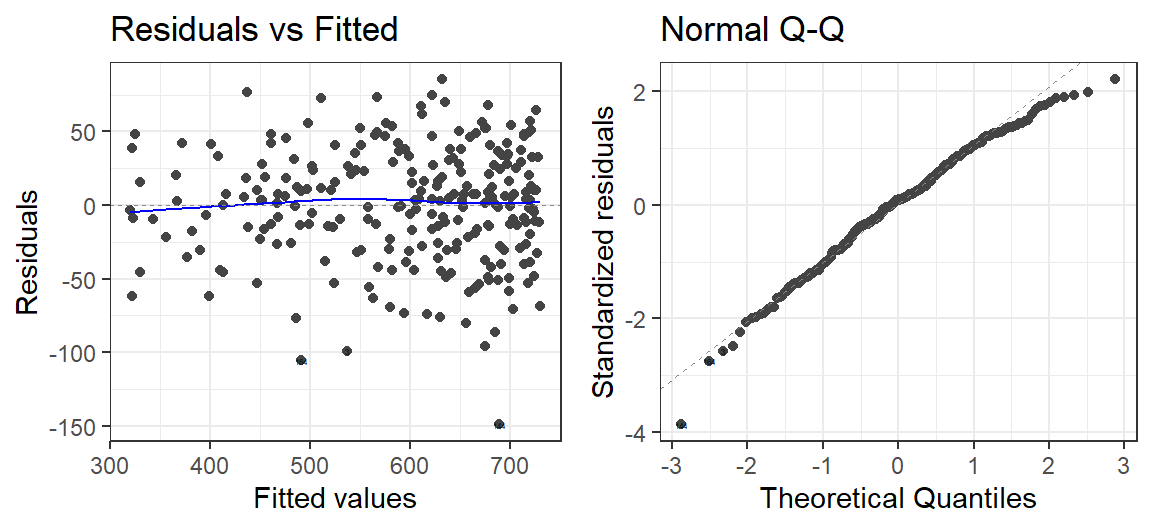
- Let’s check the residuals
autoplot(Model.Breath.Log, which = 1:2, label.size = 1) + theme_bw()
Reciprocals
- Common fix for rate data (things involving time)
- \(1/Y\)
- Adrenaline level in the blood predicted by mins after a panic attack
(mins)
- Reciprocal Example: \(Y =
(1/X)*\epsilon\)
set.seed(42)
n <- 250
# Length of panic attack
x <- 15*(runif(n, 1, 4))
y <- 1/(x)*rnorm(n,5,sd=.1)
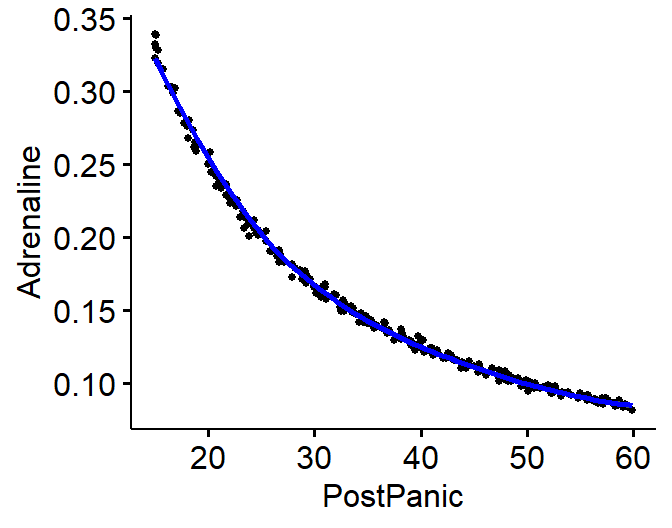
Reciprocal.Data<-data.frame(PostPanic=x,Adrenaline=y)
ggscatter(Reciprocal.Data, x = "PostPanic", y = "Adrenaline",
add = "loess", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = FALSE, # Add correlation coefficient. see ?stat_cor
size = 1 # Size of dots
)
We can take the Reciprocal of Adrenaline on the fly and replot
scatterplot(1/Adrenaline~PostPanic, data= Reciprocal.Data)
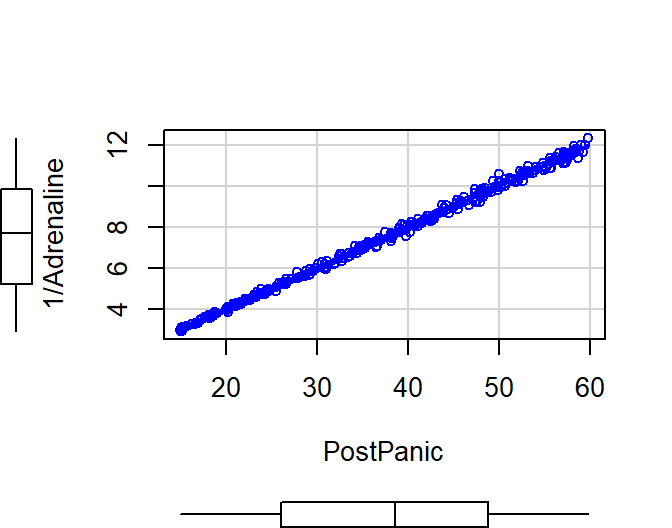
- Looks like a line now
- Let’s look at the model fit
Model.Calm.Rep<-lm(1/Adrenaline~PostPanic, data= Reciprocal.Data)
|
|
|
|
Dependent variable:
|
|
|
|
|
|
1/Adrenaline
|
|
|
|
Constant
|
-0.015 (0.029)
|
|
PostPanic
|
0.201*** (0.001)
|
|
|
|
Observations
|
250
|
|
R2
|
0.997
|
|
Adjusted R2
|
0.997
|
|
Residual Std. Error
|
0.149 (df = 248)
|
|
F Statistic
|
77,222.570*** (df = 1; 248)
|
|
|
|
Note:
|
p<0.1; p<0.05;
p<0.01
|
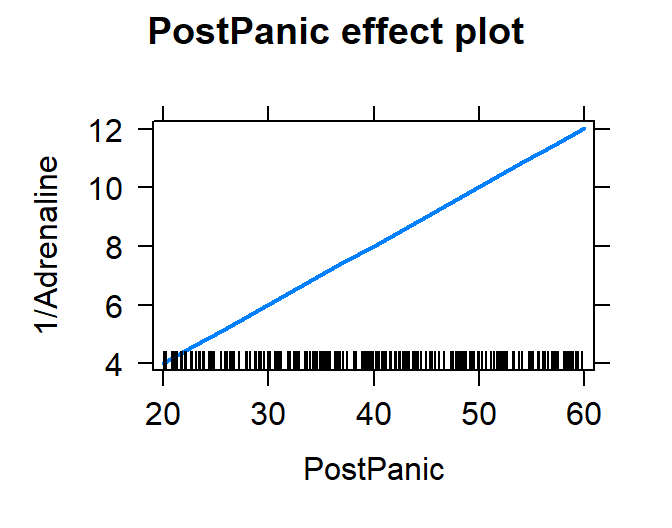
Plot the fit
plot(effect("PostPanic", Model.Calm.Rep))
Check the residuals
autoplot(Model.Calm.Rep, which = 1:2, label.size = 1) + theme_bw()
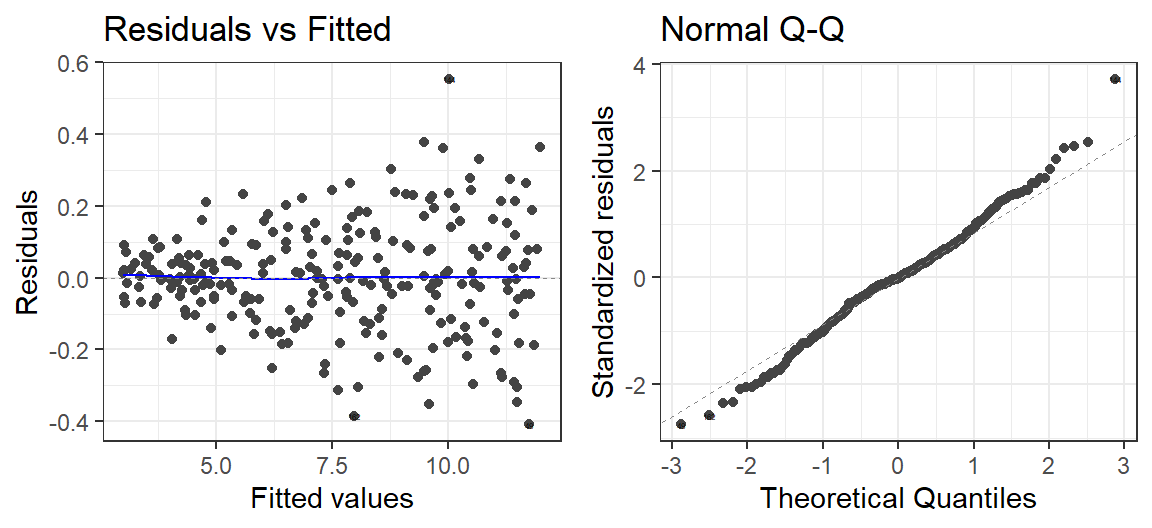
A little off (but I add in noise in a simple way)
Weird Bulges in the data (not full on curves)
- Data do not always fit our nice predefined functions
- Sometimes they clearly have bulges, but are we sure
those are real?
- Bulges can be not real in small samples or can result from some
latent factor you are not accounting for
- Let’s assume that if the data has a bulge that is real, meaningful,
and we need to account for it
- We can use the Box-Cox Transform (fits different polynomials which
we call \(\lambda\))
- \(Y = Y^\lambda - 1 / \lambda\),
where \(\lambda \neq 0\) OR \(Y = lnY\), where \(\lambda = 0\)
- In this case, we will allow \(\lambda\) to be selected based on which
makes the data most normal
- Let’s make a case with a weird power polynomial of 1.3 (something
you would not notice in your raw data)
set.seed(42)
# 250 people
n <- 250
x <- runif(n, 0, 7)
y <- -4*x^1.3+ 20*x+rnorm(n, sd=2)+20
SortaQuad.Data<-data.frame(Anxiety=x,ExamScore=y)
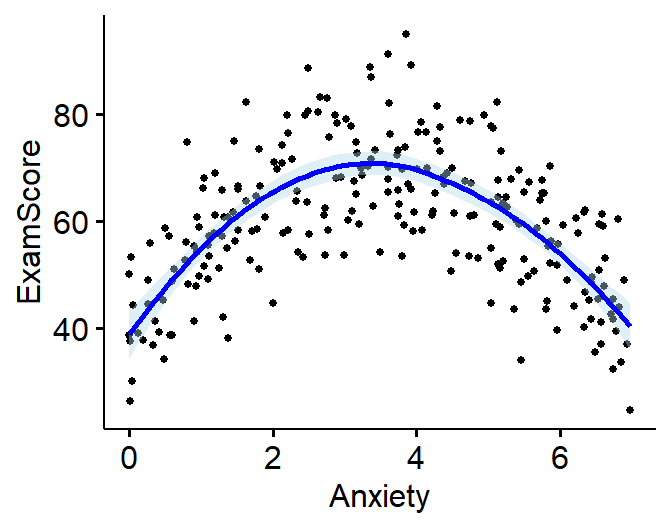
ggscatter(SortaQuad.Data, x = "Anxiety", y = "ExamScore",
add = "loess", # Add loess
add.params = list(color = "blue", fill = "lightblue"), # Customize reg. line
conf.int = TRUE, # Add confidence interval
cor.coef = FALSE, # Add correlation coefficient. see ?stat_cor
size = 1 # Size of dots
)
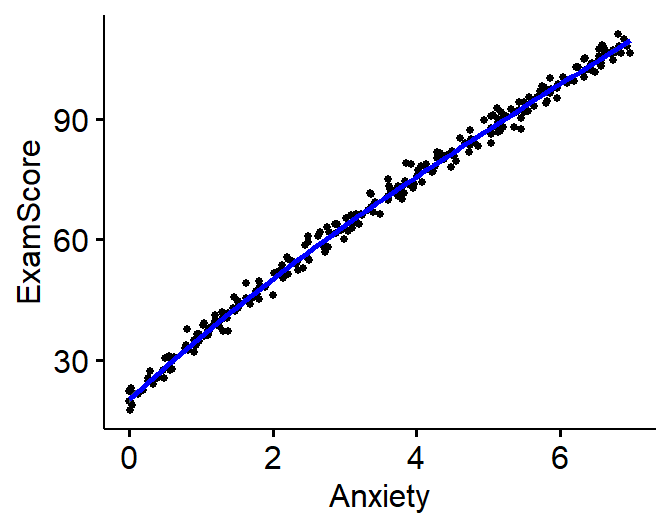
Model.SortaQuad<-lm(ExamScore~Anxiety, data= SortaQuad.Data)
autoplot(Model.SortaQuad, which = 1:2, label.size = 1) + theme_bw()
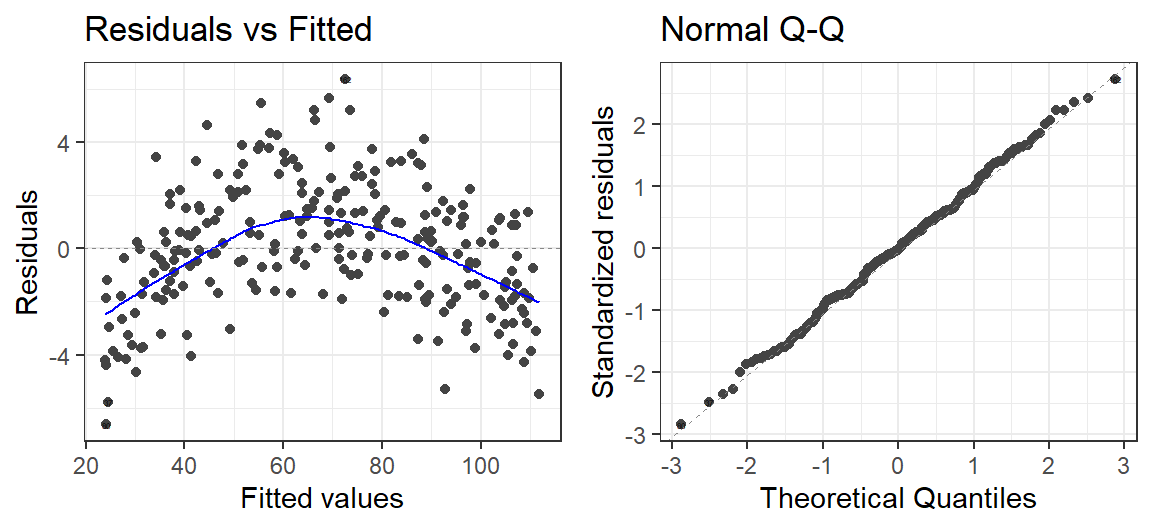
- It seems there is a slight curve in our data
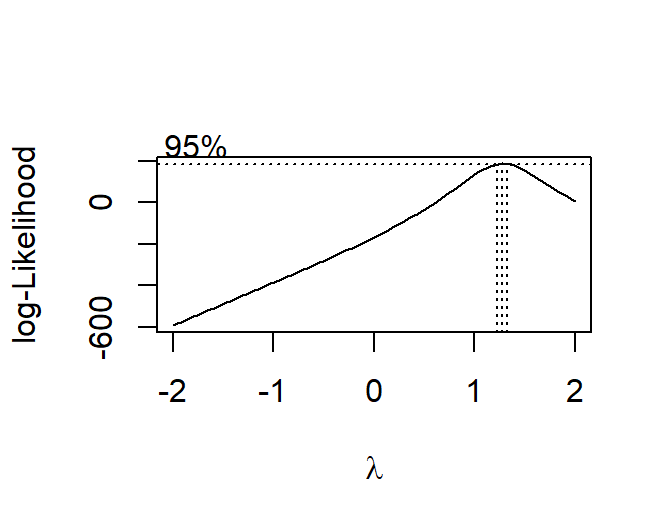
library(MASS)
bc<-boxcox(ExamScore ~Anxiety, data = SortaQuad.Data,
lambda = seq(-2, 2, len = 20))
#we can extract the lambda
(lambda <- bc$x[which.max(bc$y)])
## [1] 1.272727
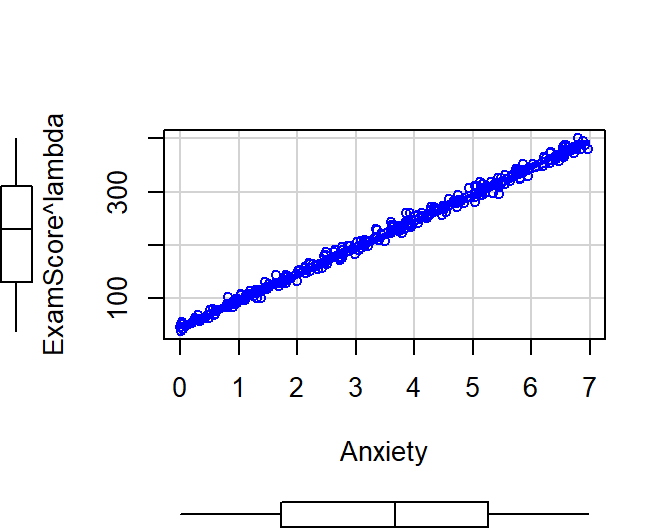
Plot the scatter and residuals from our transformed DV
scatterplot(ExamScore^lambda~Anxiety, data= SortaQuad.Data, reg.line=FALSE, smoother=loessLine)
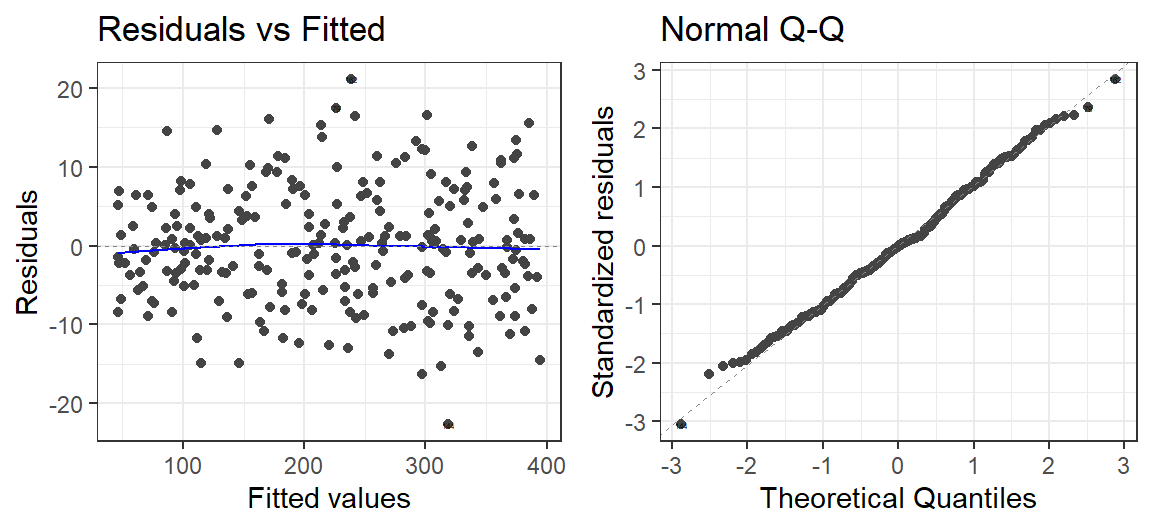
Model.SortaQuad.BC<-lm(ExamScore^lambda ~ Anxiety, data= SortaQuad.Data)
autoplot(Model.SortaQuad.BC, which = 1:2, label.size = 1) + theme_bw()
Some issues
- Box-Cox is a guess and requires some fine tuning (like adjusting the
range of lambdas)
- You have to keep track of your zeros in the DV (so you can add
constants to fix it)
- Most importantly, you have to make it harder to make sense of the DV
(but your fits will be far better using it)
LS0tDQp0aXRsZTogJ05vbi1MaW5lYXIgTW9kZWxzJw0Kb3V0cHV0Og0KICBodG1sX2RvY3VtZW50Og0KICAgIGNvZGVfZG93bmxvYWQ6IHllcw0KICAgIGZvbnRzaXplOiA4cHQNCiAgICBoaWdobGlnaHQ6IHRleHRtYXRlDQogICAgbnVtYmVyX3NlY3Rpb25zOiBubw0KICAgIHRoZW1lOiBmbGF0bHkNCiAgICB0b2M6IHllcw0KICAgIHRvY19mbG9hdDoNCiAgICAgIGNvbGxhcHNlZDogbm8NCi0tLQ0KDQpgYGB7ciBzZXR1cCwgaW5jbHVkZT1GQUxTRX0NCmtuaXRyOjpvcHRzX2NodW5rJHNldChlY2hvID0gVFJVRSkgI1Nob3cgYWxsIHNjcmlwdCBieSBkZWZhdWx0DQprbml0cjo6b3B0c19jaHVuayRzZXQobWVzc2FnZSA9IEZBTFNFKSAjaGlkZSBtZXNzYWdlcyANCmtuaXRyOjpvcHRzX2NodW5rJHNldCh3YXJuaW5nID0gIEZBTFNFKSAjaGlkZSBwYWNrYWdlIHdhcm5pbmdzIA0Ka25pdHI6Om9wdHNfY2h1bmskc2V0KGZpZy53aWR0aD0zLjUpICNTZXQgZGVmYXVsdCBmaWd1cmUgc2l6ZXMNCmtuaXRyOjpvcHRzX2NodW5rJHNldChmaWcuaGVpZ2h0PTIuNzUpICNTZXQgZGVmYXVsdCBmaWd1cmUgc2l6ZXMNCmtuaXRyOjpvcHRzX2NodW5rJHNldChmaWcuYWxpZ249J2NlbnRlcicpICNTZXQgZGVmYXVsdCBmaWd1cmUNCmtuaXRyOjpvcHRzX2NodW5rJHNldChmaWcuc2hvdyA9ICJob2xkIikgI1NldCBkZWZhdWx0IGZpZ3VyZQ0Ka25pdHI6Om9wdHNfY2h1bmskc2V0KHJlc3VsdHMgPSAiaG9sZCIpICNTZXQgZGVmYXVsdCBmaWd1cmUNCmBgYA0KDQpccGFnZWJyZWFrDQoNCiMgV2hhdCBkb2VzIExpbmVhcml0eSAmIE5vbi1MaW5lYXJpdHkgTWVhbj8NCi0gVGhlIG1ham9yaXR5IG9mIHBzeWNob2xvZ2lzdHMgb2Z0ZW4gdXNlIHRoZXNlIHRlcm1zIHRvIGRlc2NyaWJlIHRoZSByZWxhdGlvbnNoaXAgYmV0d2VlbiBZflgNCiAgICAtIExpbmVhcml0eSA9IG1hdGhlbWF0aWNhbCByZWxhdGlvbnNoaXAgYmV0d2VlbiB2YXJpYWJsZXMgY2FuIGJlIGZpdCB3aXRoIGEgKnN0cmFpZ2h0KiBsaW5lDQogICAgLSBOb24tTGluZWFyIChDdXJ2aWxpbmVhcikgPSBtYXRoZW1hdGljYWwgcmVsYXRpb25zaGlwIGJldHdlZW4gdmFyaWFibGVzIGNhbiBiZSBmaXQgd2l0aCAqbm9uLXN0cmFpZ2h0KiAoKipDdXJ2eSoqKSBsaW5lcyANCiAgICAgICAgLSBIb3dldmVyLCBiZSBjYXJlZnVsIGFzIGZvciBvdGhlciBmaWVsZHMgb3Igc3BlY2lhbHR5IGFyZWFzIG9mIHBzeWNob2xvZ3kgdGhlc2UgdGVybXMgY2FuIG1lYW4gZGlmZmVyZW50IHRoaW5nczogV2hlbiBwZW9wbGUgc2F5IG5vbi1saW5lYXIsIGFzayB0aGVtIHdoYXQgdGhleSBtZWFuDQoNCiMgTm9uLUxpbmVhciBGaXQgVHlwZXMgdGhhdCBjYW4gYmUgdHJhbnNmb3JtZWQgaW50byBsaW5lYXIgcmVncmVzc2lvbiAoYmFzaWMgdHlwZXMpDQotIFBvbHlub21pYWxzOiBQb3dlciAmIE9ydGhvZ29uYWwgDQotIEdyb3d0aDogRXhwb25lbnRpYWwsIFBvd2VyLCBMb2dhcml0aG1pYw0KLSBSYXRlczogUmVjaXByb2NhbA0KLSBDb3JyZWxhdGlvbnMNCg0KIyMgUG93ZXIgUG9seW5vbWlhbHMgDQotIE1vZGVscyB0aGF0IHNpbXBseSBjdXJ2ZXMsIHN1Y2ggYXMgcXVhZHJhdGljIG9yIGN1YmljIGVmZmVjdHMgIA0KLSBMaW5lYXI6ICRZID1CX3sxfVggKyBCXzAkDQotIFF1YWRyYXRpYzogJFkgPSBCX3sxLjJ9WCArIEJfezIuMX1YXjIgKyBCXzAkDQotIEN1YmljOiAkWSA9IEJfezEuMjN9WCArIEJfezIuMTN9WF4yICsgQl97My4xMn1YXjMgKyBCXzAkDQoNCiMjIyBTaW11bGF0ZSBhIEV4YW0gU2NvcmUgfiBBbnhpZXR5IA0KDQokWSA9IDE3LjVYIC0yLjVYXjIgKyA0MCArIFxlcHNpbG9uJA0KDQpXaGVyZSwgJFgkID0gVW5pZm9ybSBkaXN0cmlidXRpb24gb2YgTGlrZXJ0IHNjb3JlcyBiZXR3ZWVuIDAgdG8gNyAoMCA9IE5vIGFueGlldHkpICYgJFxlcHNpbG9uJCA9IFJhbmRvbSBOb3JtYWwgWyRNID0gMCQgJiAkUz0xMCRdDQoNCmBgYHtyLCBlY2hvPVRSVUUsIHdhcm5pbmc9RkFMU0V9DQpzZXQuc2VlZCg0MikJDQpuIDwtIDI1MCAjIDI1MCBwZW9wbGUNCnggPC0gcnVuaWYobiwgMCwgNykNCiMgT3VyIGVxdWF0aW9uIHRvIGNyZWF0ZSBZDQp5IDwtIC0yLjUqeF4yICsgMTcuNSp4KzQwK3Jub3JtKG4sIHNkPTEwKQ0KI0J1aWxkIG91ciBkYXRhIGZyYW1lDQpRdWFkLkRhdGE8LWRhdGEuZnJhbWUoQW54aWV0eT14LEV4YW1TY29yZT15KQ0KYGBgDQoNCiMjIFZpc3VhbGl6ZSBTaW11bGF0aW9uL0RhdGENCkJleW9uZCB2aXN1YWxpemluZyB0aGUgc2NhdHRlciBwbG90LCB3ZSBuZWVkIHNvbWUgd2F5IHRvIGVzdGltYXRlIGlmIHRoZSBlZmZlY3QgbWlnaHQgYmUgbGluZWFyIG9yIG5vbi1saW5lYXINCg0KIyMjIExvZXNzIExpbmUNCkxPRVNTID0gKipMbyoqY2FsbHkgVyoqZSoqaWdodGVkICoqUyoqY2F0dGVycGxvdCAqKlMqKm1vb3RoaW5nIA0KDQpEb2VzIGxvdHMgb2YgcmVncmVzc2lvbnMgb24gc21hbGwgc2VjdGlvbnMgb2YgdGhlIHNjYXR0ZXIgcGxvdCB0byBnZXQgdGhlIGJlc3QgZml0IChjdXJ2eSkgbGluZS4gQWxsb3dzIHlvdSB2aXN1YWxpemUgY29tcGxleCByZWxhdGlvbnNoaXBzLCBidXQgaXQgY2FuICpvdmVyZml0KiB0aGUgcmVsYXRpb25zaGlwIChzbyBiZSBjYXJlZnVsLiBUaHVzIGl0cyBhIGdvb2QgZGlhZ25vc3RpYyB0b29sLCBidXQgcmFyZWx5IHVzZWQgaW4gcGFwZXJzLg0KDQpgYGB7cn0NCmxpYnJhcnkoZ2dwdWJyKSAjZ3JhcGggZGF0YQ0KZ2dzY2F0dGVyKFF1YWQuRGF0YSwgeCA9ICJBbnhpZXR5IiwgeSA9ICJFeGFtU2NvcmUiLA0KICAgYWRkID0gImxvZXNzIiwgICMgQWRkIGxvZXNzDQogICBhZGQucGFyYW1zID0gbGlzdChjb2xvciA9ICJibHVlIiwgZmlsbCA9ICJsaWdodGJsdWUiKSwgIyBDdXN0b21pemUgcmVnLiBsaW5lDQogICBjb25mLmludCA9IFRSVUUsICMgQWRkIGNvbmZpZGVuY2UgaW50ZXJ2YWwNCiAgIGNvci5jb2VmID0gRkFMU0UsICMgQWRkIGNvcnJlbGF0aW9uIGNvZWZmaWNpZW50LiBzZWUgP3N0YXRfY29yDQogICBzaXplID0gMSAjIFNpemUgb2YgZG90cw0KICAgKQ0KYGBgDQoNCg0KIyMgRml0dGluZyBQb3dlciBQb2x5bm9taWFscyANCkZvcndhcmQgU2VsZWN0aW9uIEFwcHJvYWNoDQoNCi0gTW9kZWwgJDEkOiBMaW5lYXIgZml0DQotIE1vZGVsICQyJDogUXVhZHJhdGljIGZpdA0KLSBDaGVjayB0aGUgY2hhbmdlIGluICRSXjIkIGFuZCBzZWxlY3QgdGhlIGJlc3QgZml0IG1vZGVscw0KLSBNb2RlbCAkaSQ6IGtlZXAgZ29pbmcgdXAgdW50aWwgeW91IGFyZSBzYXRpc2ZpZWQgDQogICAgLSBOb3RlOiBIaWdoZXIgb3JkZXIgdGVybXMgKipBTFdBWVMqKiBpbmNyZWFzZSAkUl4yJCwgeW91IG5lZWQgdG8gbWFrZSBzdXJlIGl0IGlzIGJvdGggYSBzaWduaWZpY2FudCBpbXByb3ZlbWVudCBhbmQgbWVhbmluZ2Z1bCBpbXByb3ZlbWVudCANCiAgICAtIGluIFIgdGhlcmUgYXJlIHR3byB3YXlzIHRvIGFkZCBQb3dlciBQb2x5bm9taWFsczogKkkoVGVybV4yKSogb3IgdXNpbmcgdGhlICpwb2x5KiBjb21tYW5kDQoNCmBgYHtyfQ0KI0xpbmVhciBtb2RlbA0KTW9kZWwuMTwtbG0oRXhhbVNjb3JlfkFueGlldHksIGRhdGE9IFF1YWQuRGF0YSkNCiNRdWFkIG1vZGVsDQpNb2RlbC4yPC1sbShFeGFtU2NvcmV+QW54aWV0eStJKEFueGlldHleMiksIGRhdGE9IFF1YWQuRGF0YSkNCmBgYA0KDQotIFRlc3QgdG8gc2VlIGlmIHRoZSBtb2RlbCBpcyBpbXByb3ZlZCBieSB0aGUgbmV3IHRlcm0NCg0KYGBge3J9DQphbm92YShNb2RlbC4xLE1vZGVsLjIpDQpgYGANCg0KLSBFeGFtaW5lIG1vZGVsIHBhcmFtYXRlcnMNCg0KYGBge3IsIGVjaG89RkFMU0UsIHJlc3VsdHM9J2FzaXMnfQ0KbGlicmFyeShzdGFyZ2F6ZXIpDQpzdGFyZ2F6ZXIoTW9kZWwuMSxNb2RlbC4yLHR5cGU9Imh0bWwiLA0KICAgICAgICAgIGNvbHVtbi5sYWJlbHMgPSBjKCJMaW5lYXIiLCAiUXVhZHJhdGljIiksDQogICAgICAgICAgaW50ZXJjZXB0LmJvdHRvbSA9IEZBTFNFLCBzaW5nbGUucm93PVRSVUUsIA0KICAgICAgICAgIG5vdGVzLmFwcGVuZCA9IEZBTFNFLCBoZWFkZXI9RkFMU0UpDQpgYGANCg0KLSBUaGUgY2hhbmdlIGluICRSXjIkID0gYHIgc3VtbWFyeShNb2RlbC4yKSRyLnNxdWFyZSAtIHN1bW1hcnkoTW9kZWwuMSkkci5zcXVhcmVgIGFuZCB3ZSBjYW4gc2VlIHRoYXQgdGhlIGNoYW5nZSB3YXMgc2lnbmlmaWNhbnQNCi0gV291bGQgdGhlIHRoaXJkIG9yZGVyIHRlcm0gYmUgYW55IGJldHRlcj8gDQoNCmBgYHtyfQ0KI0N1YmljIG1vZGVsDQpNb2RlbC4zPC1sbShFeGFtU2NvcmV+QW54aWV0eStJKEFueGlldHleMikrSShBbnhpZXR5XjMpLCBkYXRhPSBRdWFkLkRhdGEpDQphbm92YShNb2RlbC4xLE1vZGVsLjIsTW9kZWwuMykNCmBgYA0KDQoNCmBgYHtyLCBlY2hvPUZBTFNFLHJlc3VsdHM9J2hvbGQnLHJlc3VsdHM9J2FzaXMnfQ0Kc3RhcmdhemVyKE1vZGVsLjEsTW9kZWwuMixNb2RlbC4zLHR5cGU9Imh0bWwiLA0KICAgICAgICAgIGNvbHVtbi5sYWJlbHMgPSBjKCJMaW5lYXIiLCAiUXVhZHJhdGljIiwiQ3ViaWMiKSwNCiAgICAgICAgICBpbnRlcmNlcHQuYm90dG9tID0gRkFMU0UsIHNpbmdsZS5yb3c9VFJVRSwgDQogICAgICAgICAgbm90ZXMuYXBwZW5kID0gRkFMU0UsIGhlYWRlcj1GQUxTRSkNCmBgYA0KDQoqKlNlY29uZCBvcmRlciBwb3dlciBwb2x5bm9taWFsIHdhcyBvdXIgYmVzdCBmaXQqKg0KDQojIyMgTW9kZWwgMiBpbiBEZXRhaWwNClNpbmNlIHdlIHNpbXVsYXRlZCBvdXIgbW9kZWwgd2Uga25vdyB3aGF0IHRoZSBhbnN3ZXJzICoqc2hvdWxkKiogYmUgJFkgPSAxNy41WCAtMi41WF4yICsgNDAgK1xlcHNpbG9uJA0KDQotIEV4dHJhY3Qgb3VyIHRlcm1zIGZyb20gb3VyIHJlZ3Jlc3Npb24gYW5kIGNvbXBhcmUgdG8gb3VyIHNpbXVsYXRlZCB2YWx1ZXMuDQoNCmBgYHtyfQ0KSVQ8LXJvdW5kKE1vZGVsLjIkY29lZmZpY2llbnRzWzFdLDIpDQpMVDwtcm91bmQoTW9kZWwuMiRjb2VmZmljaWVudHNbMl0sMikNClFUPC1yb3VuZChNb2RlbC4yJGNvZWZmaWNpZW50c1szXSwyKQ0KYGBgDQoNClJlZ3Jlc3Npb24gZXN0aW1hdGVkIHZhbHVlczogJFkgPSQgYHIgTFRgJFgkIGByIFFUYCRYXjIgKyQgYHIgSVRgICQrXGVwc2lsb24kDQoNCi0gT3VyIHJlZ3Jlc3Npb24gZGlkIGEgZ29vZCBqb2IsIGJ1dCB3aGF0IGlmIG91ciBzYW1wbGUgd2FzIHNtYWxsZXI/DQotIFJlZG8gb3VyIGFuYWx5c2lzIHdpdGggbiA9IDUgdG8gNjAgYW5kIHNlZSBob3cgdGhlIHRlcm1zIGNoYW5nZSBbc2VlIHNpbXVsYXRpb24gaW4gc2VwYXJhdGUgUiBmaWxlXQ0KLSB0aGUgZ3JheSBiYW5kID0gOTUlIENJDQoNCiFbXShSZWdyZXNzaW9uQ2xhc3MvU2ltcGxvdDEuanBlZykNClwNCg0KDQpJdCdzIGltcG9ydGFudCB0byBrbm93IHRoYXQgaW4gc21hbGwgc2FtcGxlcyBwb2x5bm9taWFsIGVzdGltYXRlcyBjYW4gYmUgdmVyeSB1bnN0YWJsZS4gQWxzbyBub3RlOiAqSWYgeW91IGZsaXAgdGhlIHF1YWRyYXRpYyB0ZXJtLCB0aGUgbGluZSB3aWxsIGZsaXAgZGlyZWN0aW9uKiENCg0KDQojIyMgSG93IHRvIHBsb3QgdGhpcyBmdW5jdGlvbiAoYW5kIG5vdCB0aGUgbG9lc3MgbGluZSkNCi0gWW91IGNhbiBzb2x2ZSB0aGUgcmVncmVzc2lvbiBlcXVhdGlvbiBmb3IgZGlmZmVyZW50IHZhbHVlcyBvZiB4DQotIGluIFIgeW91IGNhbiBjYWxsIHRoZSBgZWZmZWN0c2AgcGFja2FnZSB0byBkbyB3b3JrIGZvciB5b3UgdG8gc29sdmUgdGhlIHJlZ3Jlc3Npb24gZXF1YXRpb24gYXQgZGlmZmVyZW50IHZhbHVlcyBvZiAkWCQgWzAgdG8gN10uIA0KDQpgYGB7cn0NCmxpYnJhcnkoZWZmZWN0cykNCnBsb3QoZWZmZWN0KCJBbnhpZXR5IiwgTW9kZWwuMiwNCiAgICAgICAgICAgIHhsZXZlbHM9bGlzdChBbnhpZXR5PXNlcSgwLDcsMSkpKSkNCmBgYA0KDQojIyMgQ29kZSBhcHByb2FjaCB0byBwb3dlciBwb2x5bm9taWFscw0KWW91IGNhbiB1c2UgYSBjb21tYW5kIGNhbGxlZCBgcG9seWAgd2hpY2ggd2lsbCBkbyBib3RoIHBvd2VyIGFuZCBvcnRob2dvbmFsIHBvbHlub21pYWxzLiBCdXQgeW91IGNhbm5vdCBoYXZlIG1pc3NpbmcgZGF0YSB0byB1c2UgdGhpcyBjb2RlLiBUaGUgZnVuY3Rpb24gaGFzIHNlcnZhbCBpbXBvcnRhbnQgYXJndW1lbnRzIHdlIHdpbGwgZm9jdXMgb246IGBwb2x5KHgsIGRlZ3JlZSA9IDEsIHJhdyA9IEZBTFNFLCBzaW1wbGU9RkFMU0UpYC4gZGVncmVlID0gb3JkZXIgbnVtYmVyLiBTbyBgZGVncmVlID0gM2AgdGVzdHMgdGhlIGxpbmVhciwgcXVhZHJhdGljLCBhbmQgY3ViaWMuIGByYXcgPSBGQUxTRWAgdGVzdHMgdGhlIG9ydGhvZ29uYWwgdmVyc2lvbnMuIFRvIGtlZXAgdGhlbSBhcyBwb3dlciBwb2x5bm9taWFscyB3ZSBtdXN0IHNheSBgIHJhdyA9IFRSVUVgLiBMYXN0bHksIHRoZXJlIGlzIGBzaW1wbGU9RkFMU0VgLCB0aGlzIGlzIHRoZSBkZWZhdWx0IGFuZCBrZWVwcyB0aGUgYXR0cmlidXRlcyBhc3NpZ25lZCB0aGUgbmV3IHZlY3RvcnMgeW91IGhhdmUgY3JlYXRlZC4gIEkgc3VnZ2VzdCBsZWF2aW5nIHRoaXMgYWxvbmUsIG1lYW5pbmcgbGVhdmUgdGhlIGRlZmF1bHQgYXMgaXMsIGJ1dCBpdCBpcyB1c2VmdWwgdG8ga25vdyBhcyBzb21ldGltZXMgd2hlbiB3ZSBwYXNzIHRoZSByZXN1bHRzIG9mIG91ciBtb2RlbCB0aGlzIGNhbiBjcmVhdGUgcHJvYmxlbXMgd2l0aCBvdGhlciBwYWNrYWdlcyAoYnV0IEkgaGF2ZSB5ZXQgdG8gc2VlIHRoaXMpLiANCg0KYGBge3J9DQojQ3ViaWMgbW9kZWwNCk1vZGVsLjMuUDwtbG0oRXhhbVNjb3JlfnBvbHkoQW54aWV0eSwzLHJhdz1UUlVFKSwgZGF0YT0gUXVhZC5EYXRhKQ0Kc3VtbWFyeShNb2RlbC4zLlApDQpgYGANCg0KQWJvdmUgeW91IGNhbiBzZWUgdGhlIG5hbWVzIG9mIHRoZSBwYXJhbWF0ZXJzIGluIHRoZSBtb2RlbCBoYXZlIGNoYW5nZWQuIA0KDQotIHBvbHkoQW54aWV0eSwgMywgcmF3ID0gVFJVRSkxID0gbGluZWFyDQotIHBvbHkoQW54aWV0eSwgMywgcmF3ID0gVFJVRSkyID0gcXVhZHJhdGljDQotIHBvbHkoQW54aWV0eSwgMywgcmF3ID0gVFJVRSkzID0gY3ViaWMuIA0KDQojIyBPcnRob2dvbmFsIFBvbHlub21pYWxzDQpQcm9ibGVtIHdpdGggUG93ZXIgcG9seW5vbWlhbHMgaXMgaW50ZXJwcmV0aW5nIHRoZW0gaW5kZXBlbmRlbnRseS4gVGhlIGhpZ2hlciBvcmRlciB0ZXJtcyBkZXBlbmQgb24sIHRoZSBsb3dlciBvcmRlciB0ZXJtcy4gVGh1cyBsaW5lYXIgdGVybSBjYW4gYmUgc2lnbmlmaWNhbnQgY2F1c2UgdGhlIGhpZ2hlciBvcmRlciB0ZXJtIGlzIHNpZ25pZmljYW50LCBidXQgdGhlcmUgaXMgbm8gbGluZWFyIHRyZW5kIGluIHRoZSBkYXRhIChqdXN0IGxpa2UgaW4gdG9kYXkncyBleGFtcGxlKS4gKipUaGlzIGlzIGJlY2F1c2UgcG93ZXIgcG9seW5vbWlhbHMgY29ycmVsYXRlIHdpdGggZWFjaCBvdGhlciAodGhleSBhcmUgYXJlIG5vdCB1bmlxdWUgcHJlZGljdG9ycykqKg0KDQpgYGB7cn0NCkxpbmVhci5WYWx1ZXM8LWMoMSwyLDMsNCw1LDYsNykNClF1YWQuVmFsdWVzPC1MaW5lYXIuVmFsdWVzXjINCg0KTC52cy5RPC1kYXRhLmZyYW1lKExpbmVhci5WYWx1ZXM9TGluZWFyLlZhbHVlcyxRdWFkLlZhbHVlcz1RdWFkLlZhbHVlcykNCmdnc2NhdHRlcihMLnZzLlEsIHggPSAiTGluZWFyLlZhbHVlcyIsIHkgPSAiUXVhZC5WYWx1ZXMiLA0KICAgYWRkID0gInJlZy5saW5lIiwgICMgQWRkIGxvZXNzDQogICBhZGQucGFyYW1zID0gbGlzdChjb2xvciA9ICJibHVlIiwgZmlsbCA9ICJsaWdodGJsdWUiKSwgIyBDdXN0b21pemUgcmVnLiBsaW5lDQogICBjb25mLmludCA9IFRSVUUsICMgQWRkIGNvbmZpZGVuY2UgaW50ZXJ2YWwNCiAgIGNvci5jb2VmID0gVFJVRSwgIyBBZGQgY29ycmVsYXRpb24gY29lZmZpY2llbnQuIHNlZSA/c3RhdF9jb3INCiAgIHNpemUgPSAyICMgU2l6ZSBvZiBkb3RzDQogICApDQpgYGANCg0KLSBUaHVzIHRoZXkgYXJlIGJhc2ljYWxseSBtdWx0aWNvbGxpbmVhcg0KLSBMZXQncyBydW4gb3VyIFZJRiB0byB2ZXJpZnkNCg0KYGBge3J9DQpsaWJyYXJ5KGNhcikNCnZpZihNb2RlbC4yKQ0KYGBgDQoNClRlY2huaWNhbGx5LCBpdCdzIE9LIHRoYXQgdGhleSBhcmUgbXVsdGljb2xsaW5lYXIsIGJ1dCB5b3UgY2Fubm90IGludGVycHJldCBlYWNoIHRlcm0gd2l0aG91dCBsb29raW5nIGF0IHRoZSBvdGhlciAodGhlIHAtdmFsdWVzIHdpbGwgYmUgcHJvYmxlbWF0aWMpLg0KDQotIFRoZSBzb2x1dGlvbiB0byBtYWtlIHRoZSBsaW5lYXIgYW5kIHF1YWRyYXRpYyB0ZXJtcyBjb3JyZWxhdGUgd2l0aCBlYWNoIGF0IHplcm8gKGp1c3QgYXMgd2UgZGlkIHdpdGggQU5PVkEpDQotIFdlIGNhbiB1c2UgdGhlIHBvbHkgY29kZSBpbiBSIHRvIG1ha2UgdGhlbSBPcnRob2dvbmFsLiANCi0gSGVyZSBpcyBhbiBleGFtcGxlIG9mIHdoYXQgUiBpcyBnb2luZyB0byBkbzogDQoNCmBgYHtyLCBlY2hvPVRSVUUsIHdhcm5pbmc9RkFMU0V9DQpMaW5lYXJUZXJtczwtYygxLDIsMyw0LDUsNiw3KQ0KI21ha2UgdGhlbSBPcnRob2dvbmFsDQpPLlBvbHk8LXBvbHkoTGluZWFyVGVybXMsMikNCg0KTC52cy5RLk9ydGhvPC1kYXRhLmZyYW1lKExpbmVhci5WYWx1ZXMuT3J0aG89Ty5Qb2x5WywxXSxRdWFkLlZhbHVlcy5PcnRobz1PLlBvbHlbLDJdKQ0KDQpnZ3NjYXR0ZXIoTC52cy5RLk9ydGhvLCB4ID0gIkxpbmVhci5WYWx1ZXMuT3J0aG8iLCB5ID0gIlF1YWQuVmFsdWVzLk9ydGhvIiwNCiAgIGFkZCA9ICJyZWcubGluZSIsICAjIEFkZCBsb2Vzcw0KICAgYWRkLnBhcmFtcyA9IGxpc3QoY29sb3IgPSAiYmx1ZSIsIGZpbGwgPSAibGlnaHRibHVlIiksICMgQ3VzdG9taXplIHJlZy4gbGluZQ0KICAgY29uZi5pbnQgPSBUUlVFLCAjIEFkZCBjb25maWRlbmNlIGludGVydmFsDQogICBjb3IuY29lZiA9IFRSVUUsICMgQWRkIGNvcnJlbGF0aW9uIGNvZWZmaWNpZW50LiBzZWUgP3N0YXRfY29yDQogICBzaXplID0gMiAjIFNpemUgb2YgZG90cw0KICAgKQ0KYGBgDQoNCkNvcnJlbGF0aW9uIGJldHdlZW4gbGluZWFyIGFuZCBxdWFkcmF0aWMgaXMgbm93LCByID0gMCEgDQoNCmBgYHtyLCBlY2hvPVRSVUUsIHdhcm5pbmc9RkFMU0V9DQojbGV0cyBtYWtlIGEgbmV3IHBvbHkgdmVjdG9yDQpNb2RlbC4xLk88LWxtKEV4YW1TY29yZX5wb2x5KEFueGlldHksIDEpLCBkYXRhPSBRdWFkLkRhdGEpDQpNb2RlbC4yLk88LWxtKEV4YW1TY29yZX5wb2x5KEFueGlldHksIDIpLCBkYXRhPSBRdWFkLkRhdGEpDQoNCmFub3ZhKE1vZGVsLjEuTyxNb2RlbC4yLk8pDQpgYGANCg0KXHBhZ2VicmVhaw0KDQpgYGB7ciwgZWNobz1GQUxTRSwgcmVzdWx0cz0nYXNpcyd9DQpzdGFyZ2F6ZXIoTW9kZWwuMS5PLE1vZGVsLjIuTyx0eXBlPSJodG1sIiwNCiAgICAgICAgICBjb2x1bW4ubGFiZWxzID0gYygiTGluZWFyLk9ydGhvIiwgIlF1YWRyYXRpYy5PcnRobyIpLA0KICAgICAgICAgIGludGVyY2VwdC5ib3R0b20gPSBGQUxTRSwgc2luZ2xlLnJvdz1UUlVFLCANCiAgICAgICAgICBub3Rlcy5hcHBlbmQgPSBGQUxTRSwgICAgaGVhZGVyPUZBTFNFKQ0KYGBgDQoNCi0gTm90aWNlIHRoYXQgdGhlIGxpbmVhciB0ZXJtIGlzIG5vdCBzaWduaWZpY2FudCwgYnV0IHRoZSBxdWFkcmF0aWMgaXMNCiAgICAtIGJ1dCB0aGUgYWN0dWFsIGVzdGltYXRlIG9mIHRoZSBzbG9wZXMgYXJlIG5vbnNlbnNpY2FsDQotIFRoaXMgaXMgYmVjYXVzZSB5b3UgaGF2ZSAqKnJlc2NhbGVkKiogdGhlIHgtdmFsdWVzIGluIGEgZnVubnkgd2F5IChzZWUgcGxvdCBhYm92ZSkNCiAgICAtIEJ1dCBubyBtb3JlIG11bHRpY29sbGluZWFyaXR5DQotIExldCdzIHJ1biBvdXIgVklGIHRvIHZlcmlmeQ0KICAgIC0gTm90ZTogcG9seSBnZXRzIHJlYWQgdG8gdmlmIGFzIG9uZSBmYWN0b3IsIHNvIEkgd2lsbCBtYW51YWxseSBlbnRlciB0aGVzZSBpbnRvIHRoZSBtb2RlbA0KDQpgYGB7cn0NClF1YWQuRGF0YSRBbnhpZXR5Lk8uMTwtcG9seShRdWFkLkRhdGEkQW54aWV0eSwyKVssMV0NClF1YWQuRGF0YSRBbnhpZXR5Lk8uMjwtcG9seShRdWFkLkRhdGEkQW54aWV0eSwyKVssMl0NCk1vZGVsLjIuTy4yPC1sbShFeGFtU2NvcmV+QW54aWV0eS5PLjErQW54aWV0eS5PLjIsIGRhdGE9IFF1YWQuRGF0YSkNCnZpZihNb2RlbC4yLk8uMikNCmBgYA0KDQotIEFsc28gdGhlICRSXjIkIGRvIG5vdCBkaWZmZXIgYmV0d2VlbiB0aGUgcG93ZXIgb3Igb3J0aG9nb25hbCBwb2x5bm9taWFscw0KLSBPdGhlciBwcm9zIG9mIHVzaW5nIG9ydGhvZ29uYWwgcG9seW5vbWlhbHMgd2lsbCBjb21lIHdoZW4gd2Ugc3RhcnQgbG9va2luZyBhdCBpbnRlcmFjdGlvbnMNCg0KIyMjIENvZGluZyBzaG9ydGN1dCAqKFVzZSB0aGlzIGZvciB5b3VyIGFuYWx5c2lzKSoNCi0gWW91IGRvbid0IG5lZWQgdG8gcHJlbWFrZSB0aGUgdmVjdG9yIGFzIEkgZGlkIGFib3ZlDQotIFlvdSBjYW4gcHV0IHRoZSBwb2x5IGNvZGUgcmlnaHQgaW50byB0aGUgbW9kZWwNCiAgICAtIGJ1dCB0aGUgbGFiZWxpbmcgd2lsbCBjaGFuZ2UNCi0gQWxzbyBtdWNoIGVhc2llciB0byBtYWtlcyBwbG90cyBvZiB0aGUgZmluYWwgbW9kZWwgdGhpcyB3YXkNCg0KYGBge3J9DQpNb2RlbC4yLk8uUzwtbG0oRXhhbVNjb3JlfnBvbHkoQW54aWV0eSwyKSwgZGF0YT0gUXVhZC5EYXRhKQ0KYGBgDQoNCmBgYHtyLCBlY2hvPUZBTFNFLCByZXN1bHRzPSdhc2lzJ30NCnN0YXJnYXplcihNb2RlbC4yLk8uUyx0eXBlPSJodG1sIiwNCiAgICAgICAgICBjb2x1bW4ubGFiZWxzID0gYygiUXVhZHJhdGljLk9ydGhvIiksDQogICAgICAgICAgaW50ZXJjZXB0LmJvdHRvbSA9IEZBTFNFLA0KICAgICAgICAgIHNpbmdsZS5yb3c9VFJVRSwgDQogICAgICAgICAgbm90ZXMuYXBwZW5kID0gRkFMU0UsICAgICAgICAgDQogICAgICAgICAgaGVhZGVyPUZBTFNFKQ0KYGBgDQoNClRoZSBlZmZlY3RzIHBhY2thZ2Ugd2lsbCAia25vdyIgeW91IHVzZWQgYHBvbHlgIGNvbW1hbmQgYW5kIGNvbnZlcnQgdGhlIHJlc3VsdHMgYmFjayBmb3IgeW91IGF1dG9tYXRpY2FsbHkNCg0KYGBge3J9DQpwbG90KGVmZmVjdCgicG9seShBbnhpZXR5LDIpIiwgTW9kZWwuMi5PLlMsDQogICAgICAgICAgIHhsZXZlbHM9bGlzdChBbnhpZXR5PXNlcSgwLDcsMSkpKSkNCmBgYA0KDQotIEFsc28sIHlvdSBjb3VsZCBoYXZlIHNhaWQgcG9seShBbnhpZXR5LDIscmF3PVRSVUUpIHRvIGdldCBQb3dlciBwb2x5bm9taWFsIG9uIHRoZSBmbHkNCg0KIyMgRGlhZ25vc3RpY3MNCg0KLSBXaGVuIHlvdSBoYXZlIHRoZSAqKndyb25nKiogbnVtYmVyIG9mIHBvbHlub21pYWxzIHlvdSB3aWxsIGdldCBmdW5ueSBsb29raW5nIHJlc2lkdWFscw0KDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQpsaWJyYXJ5KGdnZm9ydGlmeSkNCmF1dG9wbG90KE1vZGVsLjEsIHdoaWNoID0gMToyLCBsYWJlbC5zaXplID0gMSkgKyB0aGVtZV9idygpDQpgYGANCg0KDQotIFdoZW4geW91IGhhdmUgdGhlICoqcmlnaHQqKiBudW1iZXIgb2YgcG9seW5vbWlhbHMsIHlvdSB3aWxsIGdldCBjb3JyZWN0IGxvb2tpbmcgcmVzaWR1YWxzDQotIEhlcmUgaXMgd2hlbiB3ZSB1c2UgMiB3aGVuIHdlIG5lZWQgMg0KDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQphdXRvcGxvdChNb2RlbC4yLk8uUywgd2hpY2ggPSAxOjIsIGxhYmVsLnNpemUgPSAxKSArIHRoZW1lX2J3KCkNCmBgYA0KDQpccGFnZWJyZWFrDQoNCiMgRGVhbGluZyBHcm93dGggQ3VydmVzIChQYXJ0IDIpDQotIFRoZXNlIGFyZSBtb2RlbHMgdGhhdCBjYW5ub3QgYmUgZml0IHdpdGggcG9seW5vbWlhbHMNCi0gRXhwb25lbnRpYWwgZ3Jvd3RoOiAkWSA9IGNlXntkeH0kDQotIFNldmVyaXR5IG9mIHBhbmljIGF0dGFjayAoMCB0byA1MDApIHByZWRpY3RlZCBieSBhbnhpZXR5IGxldmVsDQotIEV4cG9uZW50aWFsIEV4YW1wbGU6ICRZID0gLjA5ZV57MS4yeCtcZXBzaWxvbn0kDQotIE5vdGU6IEV1bGVyJ3MgbnVtYmVyLCAkZSA9JCBgciAgZXhwKDEpYCwgaXMgYW4gaXJyYXRpb25hbCBtYXRoIGNvbnN0YW50IGFuZCB0aGUgYmFzZSB2YWx1ZSBvZiB0aGUgbmF0dXJhbCBsb2cNCg0KYGBge3J9DQpzZXQuc2VlZCg0MikNCm4gPC0gMjUwDQp4IDwtIHJ1bmlmKG4sIDAsIDcpDQojbGV0cyBtYWtlIG91ciBlIHZhbHVlIChub3QgdGhlIGVycm9yIHRlcm0pDQplPC1leHAoMSkNCnkgPC0uMDkqZV4oMS4yMCooeCtybm9ybShuLCBzZD0uMjUpKSkNCkV4cC5EYXRhPC1kYXRhLmZyYW1lKEFueGlldHk9eCxQYW5pY0F0dGFjaz15KQ0KDQpnZ3NjYXR0ZXIoRXhwLkRhdGEsIHggPSAiQW54aWV0eSIsIHkgPSAiUGFuaWNBdHRhY2siLA0KICAgYWRkID0gImxvZXNzIiwgICMgQWRkIGxvZXNzDQogICBhZGQucGFyYW1zID0gbGlzdChjb2xvciA9ICJibHVlIiwgZmlsbCA9ICJsaWdodGJsdWUiKSwgIyBDdXN0b21pemUgcmVnLiBsaW5lDQogICBjb25mLmludCA9IFRSVUUsICMgQWRkIGNvbmZpZGVuY2UgaW50ZXJ2YWwNCiAgIGNvci5jb2VmID0gRkFMU0UsICMgQWRkIGNvcnJlbGF0aW9uIGNvZWZmaWNpZW50LiBzZWUgP3N0YXRfY29yDQogICBzaXplID0gMSAjIFNpemUgb2YgZG90cw0KICAgKQ0KYGBgDQoNCiMjIERvIFBvbHlub21pYWxzIGZhaWwgdXM/DQotIExldCdzIHRyeSBhIHNlY29uZCBvcmRlciBmaXQgc2luY2UgaXQgb25lIGN1cnZlDQpgYGB7cn0NCk1vZGVsLlBhbmljLlBvbHk8LWxtKFBhbmljQXR0YWNrfnBvbHkoQW54aWV0eSwyKSwgZGF0YT0gRXhwLkRhdGEpDQpgYGANCg0KYGBge3IsIGVjaG89RkFMU0UsIHJlc3VsdHM9J2FzaXMnfQ0Kc3RhcmdhemVyKE1vZGVsLlBhbmljLlBvbHksdHlwZT0iaHRtbCIsDQogICAgICAgICAgY29sdW1uLmxhYmVscyA9IGMoIlF1YWRyYXRpYy5PcnRobyIpLA0KICAgICAgICAgIGludGVyY2VwdC5ib3R0b20gPSBGQUxTRSwgc2luZ2xlLnJvdz1UUlVFLCANCiAgICAgICAgICBub3Rlcy5hcHBlbmQgPSBGQUxTRSwgaGVhZGVyPUZBTFNFKQ0KYGBgDQoNCi0gVXNpbmcgdGhlIHBvbHkgcmlnaHQgaW50byB0aGUgcGFja2FnZSBjYW4gYmUgdXNlZnVsIGZvciB0aGUgZWZmZWN0cyBwYWNrYWdlIGJlY2F1c2Ugd2UgY2FuIG5vdyB2aWV3IGluZGVwZW5kZW50IGltcGFjdCBvZiB0aGUgbGluZWFyIGFuZCBxdWFkcmF0aWMgdGVybXMNCg0KYGBge3J9DQpwbG90KGVmZmVjdCgicG9seShBbnhpZXR5LDIpIiwgTW9kZWwuUGFuaWMuUG9seSwNCiAgICAgICAgICAgeGxldmVscz1saXN0KEFueGlldHk9c2VxKDAsNywxKSkpKQ0KYGBgDQotIFlpa2VzLCBiYXNlZCBvbiB0aGlzIHlvdSBtaWdodCB0ZWxsIHBlb3BsZSBoYXZpbmcgemVybyBhbnhpZXR5IGlzIHdvcnNlIHRoYW4gaGF2aW5nIGxvd2VyIGFueGlldHkNCg0KLSBMZXQgY2hlY2sgdGhlIHJlc2lkdWFscw0KYGBge3IsIGZpZy53aWR0aD02fQ0KYXV0b3Bsb3QoTW9kZWwuUGFuaWMuUG9seSwgd2hpY2ggPSAxOjIsIGxhYmVsLnNpemUgPSAxKSArIHRoZW1lX2J3KCkNCmBgYA0KDQotIEFsc28gbG9va3MgcHJldHR5IG9kZA0KDQojIyBUcmFuc2Zvcm1hdGlvbnMgb2YgRXhwb25lbnRpYWwgTW9kZWwNCi0gVGhlIHNvbHV0aW9uIHRvIHRoaXMgcHJvYmxlbSBpcyBvZnRlbiB0byB0cmFuc2Zvcm0gdGhlIG9mZmVuZGluZyB2YXJpYWJsZQ0KLSBUaGUgdHlwZSBvZiB0cmFuc2Zvcm0gZGVwZW5kcyBvbiB0aGUgdHlwZSBvZiBncm93dGggbW9kZWwgeW91IGhhdmUNCi0gRXhwb25lbnRpYWwgbW9kZWw6IERlcGVuZGVudCB2YXJpYWJsZSA9IGxvZyh5KSBbaW4gciwgbG9nID0gbmF0dXJhbCBsb2ddDQotIFRodXM6ICRsb2coWSkgPSBCXzAgKyBCWCQNCi0gQW5kIHRvIGNvbnZlcnQgZnJvbSBsb2cgaW50byBtZWFuaW5nZnVsIHZhbHVlczoNCi0gJFkgPSBlXntCXzAgKyBCWH0kDQotIExldCdzIGxvb2sgYXQgTG9nKFkpIGJ5IEFueGlldHkgW3dlIHdpbGwgdXNlIHRoZSBzY2F0dGVycGxvdCBmdW5jdGlvbiBpbiBgY2FyYCBwYWNrYWdlIHRvIHNlZSB0aGlzIG1vcmUgcXVpY2tseV0NCg0KYGBge3J9DQpsaWJyYXJ5KGNhcikNCnNjYXR0ZXJwbG90KGxvZyhQYW5pY0F0dGFjayl+QW54aWV0eSwgZGF0YT0gRXhwLkRhdGEpDQpgYGANCg0KLSBMb29rcyBsaWtlIGEgbGluZSBub3cNCi0gTGV0J3MgbG9vayBhdCB0aGUgbW9kZWwgZml0DQoNCmBgYHtyfQ0KTW9kZWwuUGFuaWMuTG9nPC1sbShsb2coUGFuaWNBdHRhY2spfkFueGlldHksIGRhdGE9IEV4cC5EYXRhKQ0KYGBgDQoNCmBgYHtyLCBlY2hvPUZBTFNFLCByZXN1bHRzPSdhc2lzJ30NCk1vZGVsLlBhbmljLkxvZzwtbG0obG9nKFBhbmljQXR0YWNrKX5BbnhpZXR5LCBkYXRhPSBFeHAuRGF0YSkNCnN0YXJnYXplcihNb2RlbC5QYW5pYy5Mb2csdHlwZT0iaHRtbCIsDQogICAgICAgICAgaW50ZXJjZXB0LmJvdHRvbSA9IEZBTFNFLCBzaW5nbGUucm93PVRSVUUsIA0KICAgICAgICAgIG5vdGVzLmFwcGVuZCA9IEZBTFNFLCBoZWFkZXI9RkFMU0UpDQpgYGANCg0KUGxvdCB0aGUgZml0DQoNCmBgYHtyfQ0KcGxvdChlZmZlY3QoIkFueGlldHkiLCBNb2RlbC5QYW5pYy5Mb2csDQogICAgICAgICAgIHhsZXZlbHM9bGlzdChBbnhpZXR5PXNlcSgwLDcsMSkpKSkNCmBgYA0KDQpZb3UgY2FuIHBsb3QgaXQgbm9uLWxvZyB1bml0cyAodG8gdW5sb2cgdXNlIGV4cCBmdW5jdGlvbiksIGJ1dCB5b3UgaGF2ZSB0byB0ZWxsIHRoZSBwYWNrYWdlIHRvIGNvbnZlcnQgaXQgYmFjayB0byByYXcgc2NvcmUuDQoNCmBgYHtyLCBlY2hvPVRSVUUsbWVzc2FnZT1GQUxTRSwgd2FybmluZz1GQUxTRX0NCnBsb3QoZWZmZWN0KCJBbnhpZXR5IiwgTW9kZWwuUGFuaWMuTG9nLA0KICAgICAgICAgIHRyYW5zZm9ybWF0aW9uPWxpc3QobGluaz1sb2csIGludmVyc2U9ZXhwKSkpDQpgYGANCg0KUGxvdCB0aGUgZml0IGFuZCBjaGVjayB0aGUgcmVzaWR1YWxzDQoNCmBgYHtyLCBmaWcud2lkdGg9Nn0NCmF1dG9wbG90KE1vZGVsLlBhbmljLkxvZywgd2hpY2ggPSAxOjIsIGxhYmVsLnNpemUgPSAxKSArIHRoZW1lX2J3KCkNCmBgYA0KDQpUaG9zZSBsb29rIGFzIHRoZXkgc2hvdWxkLiANCg0KIyMjIENoYWxsZW5nZSBvZiBXb3JraW5nIHdpdGggVHJhbnNmb3Jtcw0KDQotIFByb2JsZW0gaXMgb3VyIHNjb3JlcyBhcmUgaW4gbG9nIChub3QgcmF3IHNjb3JlKQ0KLSB0byBjb252ZXJ0IGZyb20gbG9nIGludG8gbWVhbmluZ2Z1bCB2YWx1ZXM6DQotICRZID0gZV57Ql8wICsgQlh9JA0KDQpgYGB7ciwgZWNobz1UUlVFfQ0KI0ludGVyY2VwdA0KSVQuTDwtcm91bmQoTW9kZWwuUGFuaWMuTG9nJGNvZWZmaWNpZW50c1sxXSwyKQ0KI3Nsb3BlDQpMVC5MPC1yb3VuZChNb2RlbC5QYW5pYy5Mb2ckY29lZmZpY2llbnRzWzJdLDIpDQpgYGANCg0KLSAkWSA9IGUkXihgciBJVC5MYCArIGByIExULkxgWCkNCi0gRm9yIEFueGlldHkgc2NvcmUgb2YgWCA9IDAsIFkgPSBgciBleHAoMSleKElULkwgKyBMVC5MKjApYA0KLSBGb3IgQW54aWV0eSBzY29yZSBvZiBYID0gNCwgWSA9IGByIGV4cCgxKV4oSVQuTCArIExULkwqNClgDQotIEZvciBBbnhpZXR5IHNjb3JlIG9mIFggPSA3LCBZID0gYHIgZXhwKDEpXihJVC5MICsgTFQuTCo3KWANCi0gW05vdGU6IEJlIGNhcmVmdWwgYWJvdXQgd2hpY2ggbG9nIHlvdSB1c2UgKG5hdHVyYWwgb3IgYmFzZSAxMCwgeW91IGNhbiB1c2UgZWl0aGVyIGJ1dCBtYWtlIHN1cmUgdG8gc3BlY2lmeSldDQoNCiMjIFBvd2VyIGxhdw0KLSBDb21tb24gZnVuY3Rpb24gaW4gbW90b3IgY29udHJvbCBhbmQgcGVyY2VwdGlvbiBleHBlcmltZW50cw0KLSAkWSA9IGNYXmQkDQotIFNldmVyaXR5IG9mIHN3ZWF0aW5nICgwIHRvIDEwMCkgcHJlZGljdGVkIGJ5IGFueGlldHkgbGV2ZWwNCi0gRXhwb25lbnRpYWwgRXhhbXBsZTogJFkgPSAuMDcoWCleezEuN30rXGVwc2lsb24kDQotIE5vdGU6IE5hdHVyYWwgbG9nIG9mIDAgPSAgLWluZiwgc28gaWYgeW91IGhhdmUgemVyb3MgeW91IG5lZWQgdG8gYWx3YXlzIGFkZCAxDQoNCmBgYHtyLCBlY2hvPVRSVUUsIHdhcm5pbmc9RkFMU0V9DQpzZXQuc2VlZCg0MikNCm4gPC0gMjUwDQp4IDwtIHJ1bmlmKG4sIDAsIDcpKzENCnkgPC0uMDcqKHgpXjEuNw0KeSA9IHkgKiAoLjA1KiAoNStybm9ybShuLCBzZD0uMjUpKSkNClBvd2VyLkRhdGE8LWRhdGEuZnJhbWUoQW54aWV0eT14LFN3ZWF0aW5nPXkpDQoNCmdnc2NhdHRlcihQb3dlci5EYXRhLCB4ID0gIkFueGlldHkiLCB5ID0gIlN3ZWF0aW5nIiwNCiAgIGFkZCA9ICJsb2VzcyIsICAjIEFkZCBsb2Vzcw0KICAgYWRkLnBhcmFtcyA9IGxpc3QoY29sb3IgPSAiYmx1ZSIsIGZpbGwgPSAibGlnaHRibHVlIiksICMgQ3VzdG9taXplIHJlZy4gbGluZQ0KICAgY29uZi5pbnQgPSBUUlVFLCAjIEFkZCBjb25maWRlbmNlIGludGVydmFsDQogICBjb3IuY29lZiA9IEZBTFNFLCAjIEFkZCBjb3JyZWxhdGlvbiBjb2VmZmljaWVudC4gc2VlID9zdGF0X2Nvcg0KICAgc2l6ZSA9IDEgIyBTaXplIG9mIGRvdHMNCiAgICkNCmBgYA0KDQojIyMgVHJhbnNmb3JtYXRpb25zIG9mIFBvd2VyIExhdw0KLSBUaGUgc29sdXRpb24gdG8gdGhpcyBwcm9ibGVtIGlzIG9mdGVuIHRvIHRyYW5zZm9ybSB0aGUgb2ZmZW5kaW5nIHZhcmlhYmxlDQotIFBvd2VyIGxhdyBtb2RlbDogRFYgYW5kIElWIGdldCBsb2dnZWQ6IGxvZyh5KSAmIGxvZyh4KSANCi0gVGh1czogJGxvZyhZKSA9IEJfMCArIEIobG9nWCkkDQotIEFuZCB0byBjb252ZXJ0IGZyb20gbG9nIGludG8gbWVhbmluZ2Z1bCB2YWx1ZXM6DQotICRZID0gZV57Ql8wICsgQihsb2dYKX0kDQotIExldHMgbG9vayBhdCB0aGUgbG9nLWxvZyBzY2F0dGVycGxvdA0KDQpgYGB7ciwgZWNobz1UUlVFLCB3YXJuaW5nPUZBTFNFfQ0Kc2NhdHRlcnBsb3QobG9nKFN3ZWF0aW5nKX5sb2coQW54aWV0eSksIGRhdGE9IFBvd2VyLkRhdGEpDQpgYGANCg0KLSBMb29rcyBsaWtlIGEgbGluZSBub3chDQotIExldCdzIGxvb2sgYXQgdGhlIG1vZGVsIGZpdA0KDQpgYGB7cn0NCk1vZGVsLlN3ZWF0LkxMPC1sbShsb2coU3dlYXRpbmcpfmxvZyhBbnhpZXR5KSwgZGF0YT0gUG93ZXIuRGF0YSkNCmBgYA0KDQpgYGB7ciwgZWNobz1GQUxTRSxyZXN1bHRzPSdhc2lzJ30NCnN0YXJnYXplcihNb2RlbC5Td2VhdC5MTCx0eXBlPSJodG1sIiwNCiAgICAgICAgICBjb2x1bW4ubGFiZWxzID0gYygiTG9nLUxvZyBUcmFuc2Zvcm0iKSwNCiAgICAgICAgICBpbnRlcmNlcHQuYm90dG9tID0gRkFMU0UsIHNpbmdsZS5yb3c9VFJVRSwgbm90ZXMuYXBwZW5kID0gRkFMU0UsIA0KICAgICAgICAgIGhlYWRlcj1GQUxTRSkNCmBgYA0KDQpQbG90IHRoZSBmaXQgYW5kIHRlbGwgdGhlIHBhY2thZ2UgdGhhdCB5b3UgZGlkIGEgdHJhbnNmb3JtDQoNCmBgYHtyLCBlY2hvPVRSVUUsbWVzc2FnZT1GQUxTRSwgd2FybmluZz1GQUxTRX0NCnBsb3QoZWZmZWN0KCJBbnhpZXR5IiwgTW9kZWwuU3dlYXQuTEwsDQogICAgICAgICAgIHRyYW5zZm9ybWF0aW9uPWxpc3QobGluaz1sb2csIGludmVyc2U9ZXhwKSkpDQpgYGANCg0KQ2hlY2sgdGhlIHJlc2lkdWFscw0KDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQphdXRvcGxvdChNb2RlbC5Td2VhdC5MTCwgd2hpY2ggPSAxOjIsIGxhYmVsLnNpemUgPSAxKSArIHRoZW1lX2J3KCkNCmBgYA0KDQpUaGV5IGxvb2sgT0suIA0KDQojIyBMb2dhcml0aG1pYyANCi0gVGhlc2UgZnVuY3Rpb25zIGFyZSBvZnRlbiBwaHlzaWNhbCAoc291bmQgaW50ZW5zaXR5LCBlYXJ0aHF1YWtlcywgZXRjLikNCi0gJGNeWSA9IGRYJA0KLSBCcmVhdGhpbmcgcmF0ZSAoaW4gbXMgYmV0d2VlbiBicmVhdGhzKSBwcmVkaWN0ZWQgYnkgYW54aWV0eSBsZXZlbA0KLSBMb2dhcml0aG1pYyBFeGFtcGxlOiAkWSA9IC4wNShYKlxlcHNpbG9uKV57My44fSQNCg0KYGBge3J9DQpzZXQuc2VlZCg0MikNCm4gPC0gMjUwDQp4IDwtIHJ1bmlmKG4sIDAsIDcpKzENCnkgPC0yMDAqbG9nKHgqKDUrcm5vcm0obiwgc2Q9MSkpKQ0KTG9nYXJpdGhtaWMuRGF0YTwtZGF0YS5mcmFtZShBbnhpZXR5PXgsQnJlYXRoaW5nPXkpDQoNCmdnc2NhdHRlcihMb2dhcml0aG1pYy5EYXRhLCB4ID0gIkFueGlldHkiLCB5ID0gIkJyZWF0aGluZyIsDQogICBhZGQgPSAibG9lc3MiLCAgIyBBZGQgbG9lc3MNCiAgIGFkZC5wYXJhbXMgPSBsaXN0KGNvbG9yID0gImJsdWUiLCBmaWxsID0gImxpZ2h0Ymx1ZSIpLCAjIEN1c3RvbWl6ZSByZWcuIGxpbmUNCiAgIGNvbmYuaW50ID0gVFJVRSwgIyBBZGQgY29uZmlkZW5jZSBpbnRlcnZhbA0KICAgY29yLmNvZWYgPSBGQUxTRSwgIyBBZGQgY29ycmVsYXRpb24gY29lZmZpY2llbnQuIHNlZSA/c3RhdF9jb3INCiAgIHNpemUgPSAxICMgU2l6ZSBvZiBkb3RzDQogICApDQpgYGANCg0KLSBQbG90IEJyZWF0aGluZyBieSBsb2coQW54aWV0eSkNCmBgYHtyfQ0Kc2NhdHRlcnBsb3QoQnJlYXRoaW5nfmxvZyhBbnhpZXR5KSwgZGF0YT0gTG9nYXJpdGhtaWMuRGF0YSkNCmBgYA0KDQotIExvb2tzIGxpa2UgYSBsaW5lIG5vdw0KLSBMZXQncyBsb29rIGF0IHRoZSBtb2RlbCBmaXQNCg0KYGBge3J9DQpNb2RlbC5CcmVhdGguTG9nPC1sbShCcmVhdGhpbmd+bG9nKEFueGlldHkpLCBkYXRhPSBMb2dhcml0aG1pYy5EYXRhKQ0KYGBgDQpgYGB7ciwgZWNobz1GQUxTRSwgcmVzdWx0cz0nYXNpcyd9DQpzdGFyZ2F6ZXIoTW9kZWwuQnJlYXRoLkxvZyx0eXBlPSJodG1sIiwNCiAgICAgICAgICBjb2x1bW4ubGFiZWxzID0gYygiTG9nIFRyYW5zZm9ybSIpLA0KICAgICAgICAgIGludGVyY2VwdC5ib3R0b20gPSBGQUxTRSwNCiAgICAgICAgICBzaW5nbGUucm93PVRSVUUsIA0KICAgICAgICAgIG5vdGVzLmFwcGVuZCA9IEZBTFNFLCAgICAgICAgICAgDQogICAgICAgICAgaGVhZGVyPUZBTFNFKQ0KYGBgDQoNCi0gTGV0J3MgcGxvdCB0aGUgZml0IA0KYGBge3J9DQpwbG90KGVmZmVjdCgiQW54aWV0eSIsIE1vZGVsLkJyZWF0aC5Mb2cpKQ0KYGBgDQoNCi0gTGV0J3MgY2hlY2sgdGhlIHJlc2lkdWFscw0KDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQphdXRvcGxvdChNb2RlbC5CcmVhdGguTG9nLCB3aGljaCA9IDE6MiwgbGFiZWwuc2l6ZSA9IDEpICsgdGhlbWVfYncoKQ0KYGBgDQoNCiMjIFJlY2lwcm9jYWxzDQotIENvbW1vbiBmaXggZm9yIHJhdGUgZGF0YSAodGhpbmdzIGludm9sdmluZyB0aW1lKQ0KLSAkMS9ZJA0KLSBBZHJlbmFsaW5lIGxldmVsIGluIHRoZSBibG9vZCBwcmVkaWN0ZWQgYnkgbWlucyBhZnRlciBhIHBhbmljIGF0dGFjayAobWlucykNCi0gUmVjaXByb2NhbCBFeGFtcGxlOiAkWSA9ICgxL1gpKlxlcHNpbG9uJA0KDQpgYGB7cn0NCnNldC5zZWVkKDQyKQ0KbiA8LSAyNTANCiMgTGVuZ3RoIG9mIHBhbmljIGF0dGFjaw0KeCA8LSAxNSoocnVuaWYobiwgMSwgNCkpDQp5IDwtIDEvKHgpKnJub3JtKG4sNSxzZD0uMSkNClJlY2lwcm9jYWwuRGF0YTwtZGF0YS5mcmFtZShQb3N0UGFuaWM9eCxBZHJlbmFsaW5lPXkpDQoNCmdnc2NhdHRlcihSZWNpcHJvY2FsLkRhdGEsIHggPSAiUG9zdFBhbmljIiwgeSA9ICJBZHJlbmFsaW5lIiwNCiAgIGFkZCA9ICJsb2VzcyIsICAjIEFkZCBsb2Vzcw0KICAgYWRkLnBhcmFtcyA9IGxpc3QoY29sb3IgPSAiYmx1ZSIsIGZpbGwgPSAibGlnaHRibHVlIiksICMgQ3VzdG9taXplIHJlZy4gbGluZQ0KICAgY29uZi5pbnQgPSBUUlVFLCAjIEFkZCBjb25maWRlbmNlIGludGVydmFsDQogICBjb3IuY29lZiA9IEZBTFNFLCAjIEFkZCBjb3JyZWxhdGlvbiBjb2VmZmljaWVudC4gc2VlID9zdGF0X2Nvcg0KICAgc2l6ZSA9IDEgIyBTaXplIG9mIGRvdHMNCiAgICkNCmBgYA0KDQpXZSBjYW4gdGFrZSB0aGUgUmVjaXByb2NhbCBvZiBBZHJlbmFsaW5lIG9uIHRoZSBmbHkgYW5kIHJlcGxvdA0KDQpgYGB7cn0NCnNjYXR0ZXJwbG90KDEvQWRyZW5hbGluZX5Qb3N0UGFuaWMsIGRhdGE9IFJlY2lwcm9jYWwuRGF0YSkNCmBgYA0KDQotIExvb2tzIGxpa2UgYSBsaW5lIG5vdw0KLSBMZXQncyBsb29rIGF0IHRoZSBtb2RlbCBmaXQNCg0KYGBge3J9DQpNb2RlbC5DYWxtLlJlcDwtbG0oMS9BZHJlbmFsaW5lflBvc3RQYW5pYywgZGF0YT0gUmVjaXByb2NhbC5EYXRhKQ0KYGBgDQoNCmBgYHtyLCBlY2hvPUZBTFNFLHJlc3VsdHM9J2FzaXMnfQ0Kc3RhcmdhemVyKE1vZGVsLkNhbG0uUmVwLHR5cGU9Imh0bWwiLA0KICAgICAgICAgIGludGVyY2VwdC5ib3R0b20gPSBGQUxTRSwNCiAgICAgICAgICBzaW5nbGUucm93PVRSVUUsIA0KICAgICAgICAgIG5vdGVzLmFwcGVuZCA9IEZBTFNFLCAgICAgICAgICANCiAgICAgICAgICBoZWFkZXI9RkFMU0UpDQpgYGANCg0KUGxvdCB0aGUgZml0IA0KYGBge3IsIGVjaG89VFJVRSxtZXNzYWdlPUZBTFNFLCB3YXJuaW5nPUZBTFNFfQ0KcGxvdChlZmZlY3QoIlBvc3RQYW5pYyIsIE1vZGVsLkNhbG0uUmVwKSkNCmBgYA0KDQpDaGVjayB0aGUgcmVzaWR1YWxzDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQphdXRvcGxvdChNb2RlbC5DYWxtLlJlcCwgd2hpY2ggPSAxOjIsIGxhYmVsLnNpemUgPSAxKSArIHRoZW1lX2J3KCkNCmBgYA0KDQpBIGxpdHRsZSBvZmYgKGJ1dCBJIGFkZCBpbiBub2lzZSBpbiBhIHNpbXBsZSB3YXkpDQoNCiMjIFdlaXJkIEJ1bGdlcyBpbiB0aGUgZGF0YSAobm90IGZ1bGwgb24gY3VydmVzKQ0KLSBEYXRhIGRvIG5vdCBhbHdheXMgZml0IG91ciBuaWNlIHByZWRlZmluZWQgZnVuY3Rpb25zDQotIFNvbWV0aW1lcyB0aGV5IGNsZWFybHkgaGF2ZSAqKmJ1bGdlcyoqLCBidXQgYXJlIHdlIHN1cmUgdGhvc2UgYXJlIHJlYWw/DQotIEJ1bGdlcyBjYW4gYmUgbm90IHJlYWwgaW4gc21hbGwgc2FtcGxlcyBvciBjYW4gcmVzdWx0IGZyb20gc29tZSBsYXRlbnQgZmFjdG9yIHlvdSBhcmUgbm90IGFjY291bnRpbmcgZm9yDQotIExldCdzIGFzc3VtZSB0aGF0IGlmIHRoZSBkYXRhIGhhcyBhIGJ1bGdlIHRoYXQgaXMgcmVhbCwgbWVhbmluZ2Z1bCwgYW5kIHdlIG5lZWQgdG8gYWNjb3VudCBmb3IgaXQNCi0gV2UgY2FuIHVzZSB0aGUgQm94LUNveCBUcmFuc2Zvcm0gKGZpdHMgZGlmZmVyZW50IHBvbHlub21pYWxzIHdoaWNoIHdlIGNhbGwgJFxsYW1iZGEkKQ0KLSAkWSA9IFleXGxhbWJkYSAtIDEgLyBcbGFtYmRhJCwgd2hlcmUgJFxsYW1iZGEgXG5lcSAwJCBPUiAkWSA9IGxuWSQsIHdoZXJlICRcbGFtYmRhID0gMCQNCi0gSW4gdGhpcyBjYXNlLCB3ZSB3aWxsIGFsbG93ICRcbGFtYmRhJCB0byBiZSBzZWxlY3RlZCBiYXNlZCBvbiB3aGljaCBtYWtlcyB0aGUgZGF0YSBtb3N0IG5vcm1hbA0KLSBMZXQncyBtYWtlIGEgY2FzZSB3aXRoIGEgd2VpcmQgcG93ZXIgcG9seW5vbWlhbCBvZiAxLjMgKHNvbWV0aGluZyB5b3Ugd291bGQgbm90IG5vdGljZSBpbiB5b3VyIHJhdyBkYXRhKQ0KDQpgYGB7cn0NCnNldC5zZWVkKDQyKQ0KIyAyNTAgcGVvcGxlDQpuIDwtIDI1MA0KeCA8LSBydW5pZihuLCAwLCA3KQ0KeSA8LSAtNCp4XjEuMysgMjAqeCtybm9ybShuLCBzZD0yKSsyMA0KU29ydGFRdWFkLkRhdGE8LWRhdGEuZnJhbWUoQW54aWV0eT14LEV4YW1TY29yZT15KQ0KDQpnZ3NjYXR0ZXIoU29ydGFRdWFkLkRhdGEsIHggPSAiQW54aWV0eSIsIHkgPSAiRXhhbVNjb3JlIiwNCiAgIGFkZCA9ICJsb2VzcyIsICAjIEFkZCBsb2Vzcw0KICAgYWRkLnBhcmFtcyA9IGxpc3QoY29sb3IgPSAiYmx1ZSIsIGZpbGwgPSAibGlnaHRibHVlIiksICMgQ3VzdG9taXplIHJlZy4gbGluZQ0KICAgY29uZi5pbnQgPSBUUlVFLCAjIEFkZCBjb25maWRlbmNlIGludGVydmFsDQogICBjb3IuY29lZiA9IEZBTFNFLCAjIEFkZCBjb3JyZWxhdGlvbiBjb2VmZmljaWVudC4gc2VlID9zdGF0X2Nvcg0KICAgc2l6ZSA9IDEgIyBTaXplIG9mIGRvdHMNCiAgICkNCmBgYA0KDQotIExldCdzIHRyeSBhIGxpbmVhciBtb2RlbA0KDQpgYGB7cn0NCk1vZGVsLlNvcnRhUXVhZDwtbG0oRXhhbVNjb3JlfkFueGlldHksIGRhdGE9IFNvcnRhUXVhZC5EYXRhKQ0KYGBgDQotIFBsb3QgdGhlIFJlc2lkdWFscw0KDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQphdXRvcGxvdChNb2RlbC5Tb3J0YVF1YWQsIHdoaWNoID0gMToyLCBsYWJlbC5zaXplID0gMSkgKyB0aGVtZV9idygpDQpgYGANCg0KLSBJdCBzZWVtcyB0aGVyZSBpcyBhIHNsaWdodCBjdXJ2ZSBpbiBvdXIgZGF0YQ0KDQpgYGB7cn0NCmxpYnJhcnkoTUFTUykNCmJjPC1ib3hjb3goRXhhbVNjb3JlIH5BbnhpZXR5LCBkYXRhID0gU29ydGFRdWFkLkRhdGEsDQogICAgICAgbGFtYmRhID0gc2VxKC0yLCAyLCBsZW4gPSAyMCkpDQojd2UgY2FuIGV4dHJhY3QgdGhlIGxhbWJkYQ0KKGxhbWJkYSA8LSBiYyR4W3doaWNoLm1heChiYyR5KV0pDQpgYGANCg0KUGxvdCB0aGUgc2NhdHRlciBhbmQgcmVzaWR1YWxzIGZyb20gb3VyIHRyYW5zZm9ybWVkIERWDQpgYGB7cn0NCnNjYXR0ZXJwbG90KEV4YW1TY29yZV5sYW1iZGF+QW54aWV0eSwgZGF0YT0gU29ydGFRdWFkLkRhdGEsIHJlZy5saW5lPUZBTFNFLCBzbW9vdGhlcj1sb2Vzc0xpbmUpDQpNb2RlbC5Tb3J0YVF1YWQuQkM8LWxtKEV4YW1TY29yZV5sYW1iZGEgfiBBbnhpZXR5LCBkYXRhPSBTb3J0YVF1YWQuRGF0YSkNCmBgYA0KDQpgYGB7ciwgZmlnLndpZHRoPTZ9DQphdXRvcGxvdChNb2RlbC5Tb3J0YVF1YWQuQkMsIHdoaWNoID0gMToyLCBsYWJlbC5zaXplID0gMSkgKyB0aGVtZV9idygpDQpgYGANCg0KIyMjIFNvbWUgaXNzdWVzDQotIEJveC1Db3ggaXMgYSBndWVzcyBhbmQgcmVxdWlyZXMgc29tZSBmaW5lIHR1bmluZyAobGlrZSBhZGp1c3RpbmcgdGhlIHJhbmdlIG9mIGxhbWJkYXMpDQotIFlvdSBoYXZlIHRvIGtlZXAgdHJhY2sgb2YgeW91ciB6ZXJvcyBpbiB0aGUgRFYgKHNvIHlvdSBjYW4gYWRkIGNvbnN0YW50cyB0byBmaXggaXQpDQotIE1vc3QgaW1wb3J0YW50bHksIHlvdSBoYXZlIHRvIG1ha2UgaXQgaGFyZGVyIHRvIG1ha2Ugc2Vuc2Ugb2YgdGhlIERWIChidXQgeW91ciBmaXRzIHdpbGwgYmUgZmFyIGJldHRlciB1c2luZyBpdCkNCg0KPHNjcmlwdD4NCiAgKGZ1bmN0aW9uKGkscyxvLGcscixhLG0pe2lbJ0dvb2dsZUFuYWx5dGljc09iamVjdCddPXI7aVtyXT1pW3JdfHxmdW5jdGlvbigpew0KICAoaVtyXS5xPWlbcl0ucXx8W10pLnB1c2goYXJndW1lbnRzKX0saVtyXS5sPTEqbmV3IERhdGUoKTthPXMuY3JlYXRlRWxlbWVudChvKSwNCiAgbT1zLmdldEVsZW1lbnRzQnlUYWdOYW1lKG8pWzBdO2EuYXN5bmM9MTthLnNyYz1nO20ucGFyZW50Tm9kZS5pbnNlcnRCZWZvcmUoYSxtKQ0KICB9KSh3aW5kb3csZG9jdW1lbnQsJ3NjcmlwdCcsJ2h0dHBzOi8vd3d3Lmdvb2dsZS1hbmFseXRpY3MuY29tL2FuYWx5dGljcy5qcycsJ2dhJyk7DQoNCiAgZ2EoJ2NyZWF0ZScsICdVQS05MDQxNTE2MC0xJywgJ2F1dG8nKTsNCiAgZ2EoJ3NlbmQnLCAncGFnZXZpZXcnKTsNCg0KPC9zY3JpcHQ+DQoNCg==









 - Yikes, based on this you might tell people having zero anxiety is
worse than having lower anxiety
- Yikes, based on this you might tell people having zero anxiety is
worse than having lower anxiety