Subjects & Items: Parsimonious Modeling
Maximal Modeling
- Last week we reviewed why Barr et al., 2013 proposed to reduce the
type I error rate and increase the power using the maximal random
structure:
lmer(DV~ A*B + (1+A*B:Subjects) + (1+A*B:Items)) - However, we ran into convergence issue in practice and seemed like trial and error was needed to find the best random structure combination. Further, the more within-subject factors you have, the more complex you random structure become and harder it is to reduce.
Parsimonious Approach
Bates et al. (2015) [RePsychLing package] &
Matuschek et al. (2017) provide a principled approach to finding the
best random structure for your data. Bates et al. suggest the
convergence issues are because the random effects are too complex for
the actual data (and overparameterized non-convergent LMM are not
interpretable). They provide the example of in a 2x2 within-subject
design you will have [(2X2 subjects) + (2X2 items) + 1 / 2] = 10 random
parameters. When you go to 2x2x2, we will get 36 random parameters. A
warning sign that your model is overparameterized are high random
correlations, \(r > [-.8 ,
.8]\).
They suggest conducting a PCA analysis of the random effects (of the maximal model) to determine how random components are really needed explaining and if we need all the parameters we are using (like in factor analysis we can examine a scree plot). Even if the maximal model converges does not mean we need all those random parameters. The benefit of pruning the random effects back allows us to protect our Type I error but increase our power (Matuschek et al., 2017).
Once we figure out the most complex structure the data can sustain, we can use model fit comparisons of nested models to find the best random effects.
Simulation
- We will simulate a full crossed study with 30 subjects and 10 items, but we will examine results were layers levels of random effects to see how parsimonious modeling works.
: Fully crossed:
(1+C1*C2|Subject) + (1+C1*C2|Item) + (1|Subject:Item)
- we will ignore the
(1|Subject:Item)for simplicity today - Simulations generated in Excel File we reviewed at the start of class.
- 2 x 2 repeated measure with 10 items: Population Effect: \(Y = 4X_1-4X_2+4X_1X_2+10\)
- [Simulation results in
Sim8.csv] - Conditions are effects coded \((-.5, .5)\)
- Download Data
DataSim8<-read.csv("Mixed/Sim8.csv")
DataSim8$C1<-factor(DataSim8$Condition1)
DataSim8$C2<-as.factor(DataSim8$Condition2)
DataSim8$Item<-as.factor(DataSim8$Item)
DataSim8$Subject<-as.factor(DataSim8$Subject)Simulation 1: No Random Slopes/Items
- Here we simulate population effects for our 2x2 design, but for each subject we simply add noise to each measurement \(Y = 4X_1-4X_2+4X_1X_2+10 + \epsilon\) . Thus there are No random slopes per subject, and there are no item level effects. In short, this is basically a plain old linear regression, and every measurement per subject is independent (an lm model would be fine with this data).
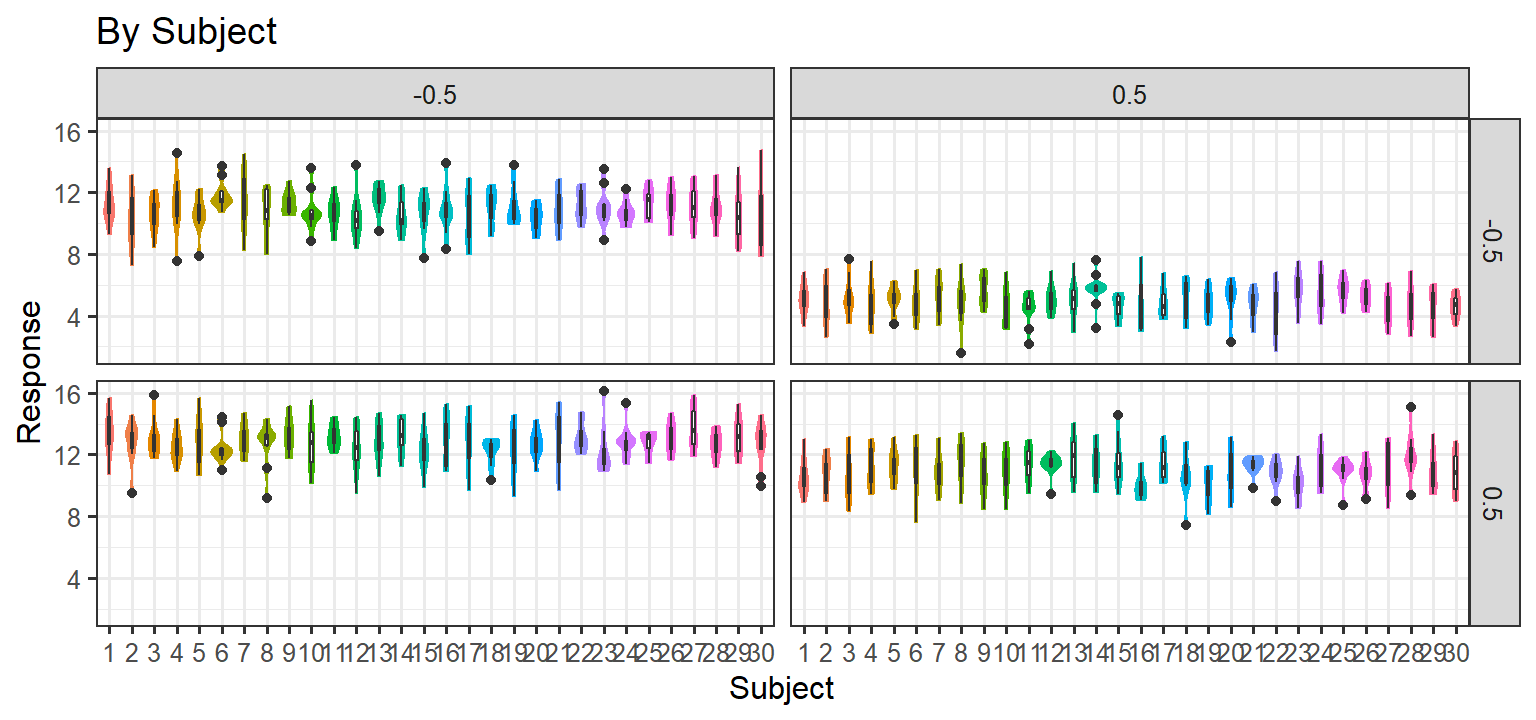
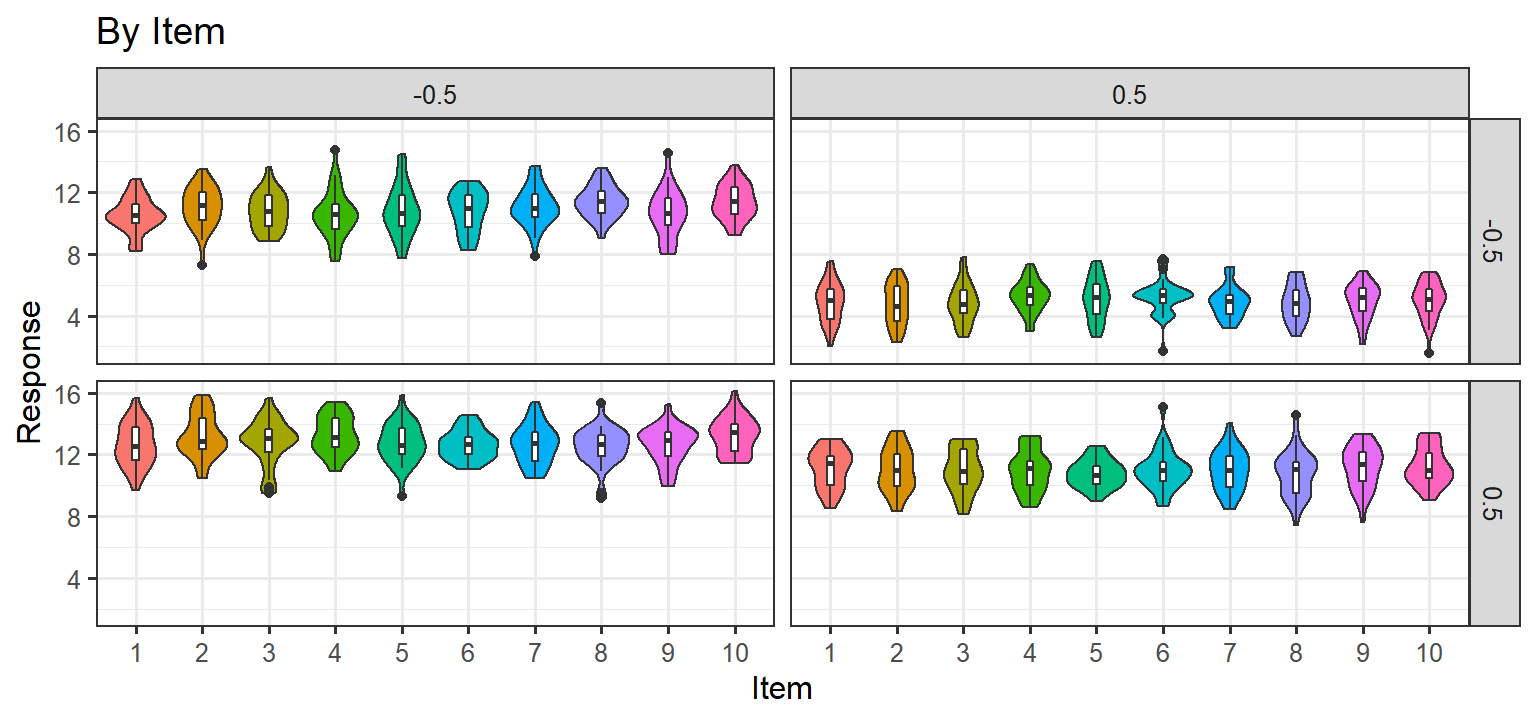
Parsimonious Stages
- Step 1:
(1+C1*C2|Subject) + (1+C1*C2|Item)
library(lme4)
FullMax.SSNoise<-lmer(DV_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1+Condition1*Condition2|Item),
data=DataSim8, REML=FALSE)
summary(FullMax.SSNoise, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 |
## Subject) + (1 + Condition1 * Condition2 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4045.5 4172.7 -1997.7 3995.5 1175
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.95067 -0.66431 0.01886 0.66489 3.13739
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 0.000212 0.01456
## Condition1 0.059057 0.24302 1.00
## Condition2 0.006627 0.08141 1.00 1.00
## Condition1:Condition2 0.068101 0.26096 1.00 1.00 1.00
## Item (Intercept) 0.004994 0.07066
## Condition1 0.012173 0.11033 0.44
## Condition2 0.024671 0.15707 -0.83 0.13
## Condition1:Condition2 0.062291 0.24958 -0.09 -0.94 -0.47
## Residual 1.604201 1.26657
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 9.93133 0.04293 15.31430 231.32 < 2e-16 ***
## Condition1 3.97857 0.09238 18.67659 43.07 < 2e-16 ***
## Condition2 -3.88643 0.08964 13.92792 -43.36 2.93e-16 ***
## Condition1:Condition2 4.09045 0.17288 16.22527 23.66 5.24e-14 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- The model failed to converge and problems with our correlations and zero intercepts
- Step 2: extract the PCA of the random structure from the maximal model
library(RePsychLing)
FullMax.SSNoise.PCA<-rePCA(FullMax.SSNoise)
summary(FullMax.SSNoise.PCA)## $Subject
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 0.289 0.0007822 5.401e-05 3.246e-06
## Proportion of Variance 1.000 0.0000100 0.000e+00 0.000e+00
## Cumulative Proportion 1.000 1.0000000 1.000e+00 1.000e+00
##
## $Item
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 0.2221 0.1247 4.451e-05 1.171e-05
## Proportion of Variance 0.7603 0.2397 0.000e+00 0.000e+00
## Cumulative Proportion 0.7603 1.0000 1.000e+00 1.000e+00- For
Subject: we see the first component captures 100% of the random variance. - For
Item: we see the first two components capture 100% of the random variance. - Step 3: Reduce complexity Cycle:
Cycle 1
- The random correlations are showing problems, lets remove them.
FullMax.SSNoise.2<-lmer(DV_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2||Subject)
+(1+Condition1*Condition2||Item),
data=DataSim8, REML=FALSE)
summary(FullMax.SSNoise.2, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula:
## DV_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 ||
## Subject) + (1 + Condition1 * Condition2 || Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4026.2 4092.4 -2000.1 4000.2 1187
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.93808 -0.67371 0.01669 0.64739 3.14696
##
## Random effects:
## Groups Name Variance Std.Dev.
## Subject (Intercept) 0.000e+00 0.000e+00
## Subject.1 Condition1 4.845e-02 2.201e-01
## Subject.2 Condition2 7.051e-10 2.655e-05
## Subject.3 Condition1:Condition2 9.043e-10 3.007e-05
## Item (Intercept) 2.898e-04 1.702e-02
## Item.1 Condition1 0.000e+00 0.000e+00
## Item.2 Condition2 8.432e-03 9.182e-02
## Item.3 Condition1:Condition2 2.042e-03 4.519e-02
## Residual 1.629e+00 1.276e+00
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 9.93133 0.03723 10.00456 266.76 < 2e-16 ***
## Condition1 3.97857 0.08392 29.99973 47.41 < 2e-16 ***
## Condition2 -3.88643 0.07919 9.99973 -49.08 2.98e-13 ***
## Condition1:Condition2 4.09045 0.14805 9.97458 27.63 9.34e-11 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- Recheck the PCA
FullMax.SSNoise.PCA2<-rePCA(FullMax.SSNoise.2)
summary(FullMax.SSNoise.PCA2)## $Subject
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 0.1725 2.356e-05 2.081e-05 0
## Proportion of Variance 1.0000 0.000e+00 0.000e+00 0
## Cumulative Proportion 1.0000 1.000e+00 1.000e+00 1
##
## $Item
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 0.07195 0.03541 0.01334 0
## Proportion of Variance 0.78338 0.18970 0.02692 0
## Cumulative Proportion 0.78338 0.97308 1.00000 1- Model is still overparameterized.
Cycle 2
- Remove the zero random effect
FullMax.SSNoise.3<-lmer(DV_SSNoise ~ Condition1*Condition2
+(1+Condition1||Subject)
+(1+Condition2||Item),
data=DataSim8, REML=FALSE)
summary(FullMax.SSNoise.3, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 || Subject) +
## (1 + Condition2 || Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4018.2 4064.0 -2000.1 4000.2 1191
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.93921 -0.67450 0.01678 0.64702 3.14649
##
## Random effects:
## Groups Name Variance Std.Dev.
## Subject (Intercept) 0.0000000 0.00000
## Subject.1 Condition1 0.0484452 0.22010
## Item (Intercept) 0.0002885 0.01698
## Item.1 Condition2 0.0084285 0.09181
## Residual 1.6286530 1.27619
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 9.93133 0.03723 10.00135 266.76 < 2e-16 ***
## Condition1 3.97857 0.08393 30.00000 47.41 < 2e-16 ***
## Condition2 -3.88643 0.07919 10.00007 -49.08 2.98e-13 ***
## Condition1:Condition2 4.09045 0.14736 1150.00164 27.76 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- Recheck the PCA
FullMax.SSNoise.PCA3<-rePCA(FullMax.SSNoise.3)
summary(FullMax.SSNoise.PCA3)## $Subject
## Importance of components:
## [,1] [,2]
## Standard deviation 0.1725 0
## Proportion of Variance 1.0000 0
## Cumulative Proportion 1.0000 1
##
## $Item
## Importance of components:
## [,1] [,2]
## Standard deviation 0.07194 0.01331
## Proportion of Variance 0.96691 0.03309
## Cumulative Proportion 0.96691 1.00000- Model is still way overparameterized.
Cycle 3
- Clear we don’t need the random slope on the subject.
- Given that random slope on items captures .03% of the variance we may not need it.
FullMax.SSNoise.4<-lmer(DV_SSNoise ~ Condition1*Condition2
+(1|Subject)
+(1|Item),
data=DataSim8, REML=FALSE)
summary(FullMax.SSNoise.4, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SSNoise ~ Condition1 * Condition2 + (1 | Subject) + (1 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4015.4 4051.0 -2000.7 4001.4 1193
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.8825 -0.6529 0.0059 0.6545 3.1948
##
## Random effects:
## Groups Name Variance Std.Dev.
## Subject (Intercept) 0.0000000 0.00000
## Item (Intercept) 0.0001682 0.01297
## Residual 1.6429917 1.28179
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 9.93133 0.03723 9.97956 266.76 <2e-16 ***
## Condition1 3.97857 0.07400 1189.97847 53.76 <2e-16 ***
## Condition2 -3.88643 0.07400 1189.97847 -52.52 <2e-16 ***
## Condition1:Condition2 4.09045 0.14801 1189.97847 27.64 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- Check fit
## Data: DataSim8
## Models:
## FullMax.SSNoise.4: DV_SSNoise ~ Condition1 * Condition2 + (1 | Subject) + (1 | Item)
## FullMax.SSNoise.3: DV_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 || Subject) + (1 + Condition2 || Item)
## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
## FullMax.SSNoise.4 7 4015.4 4051 -2000.7 4001.4
## FullMax.SSNoise.3 9 4018.2 4064 -2000.1 4000.2 1.175 2 0.5557Results of Simulation 1
Random Intercepts model is as good a fit as that last overparameterized model. As would expect, there are no random slopes or item level effects (LMM is not needed, but the process told us that pretty clearly)
Simulation 2: Random Slopes per subject + Noise/No Item level effect
- Here we let each subject have their own slope for each condition (1
& 2) and the interaction. We also add noise to each trial, but items
have no random effect. Thus there are ARE random slopes per subject, and
there are no item level effects. If this approach matches the data, we
will be left with this random structure:
(1+C1*C2|Subject) + (1|Item)
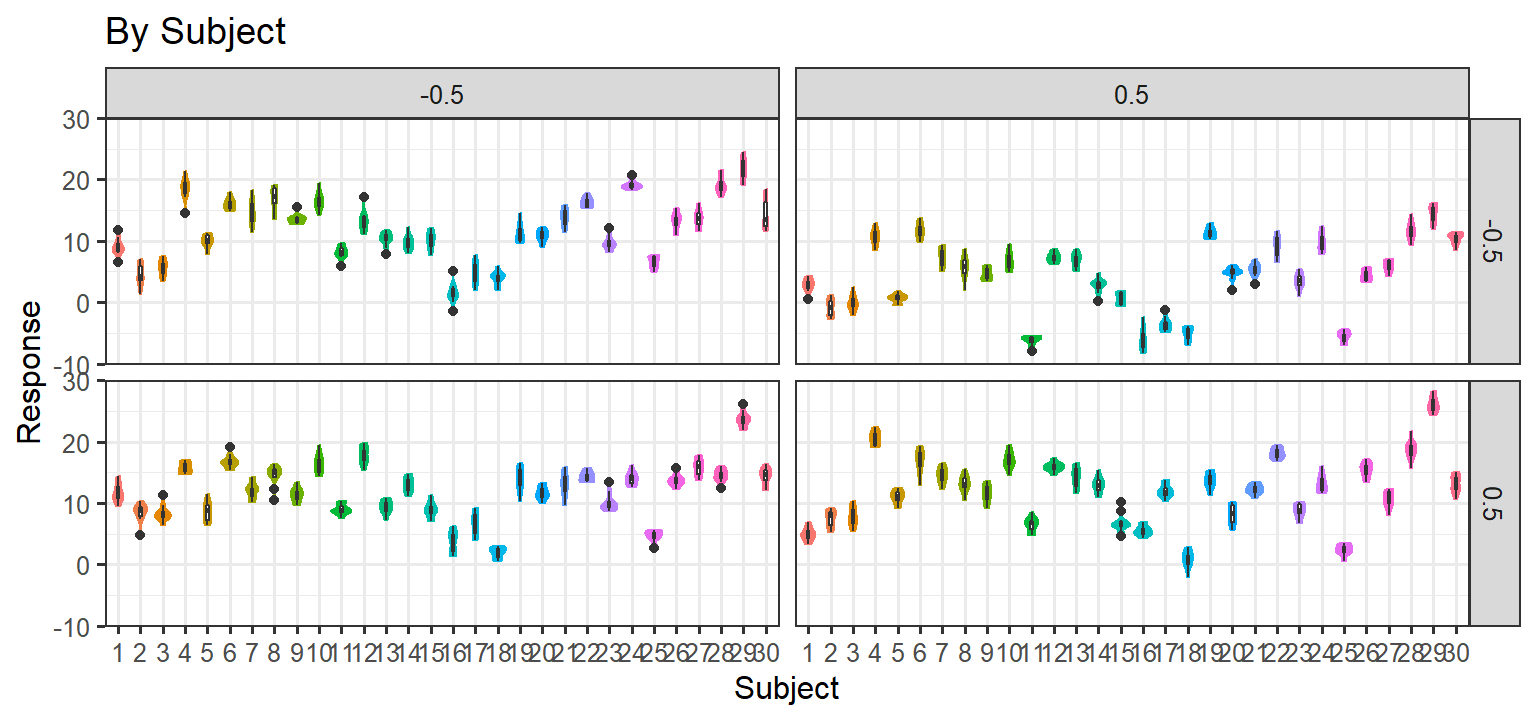
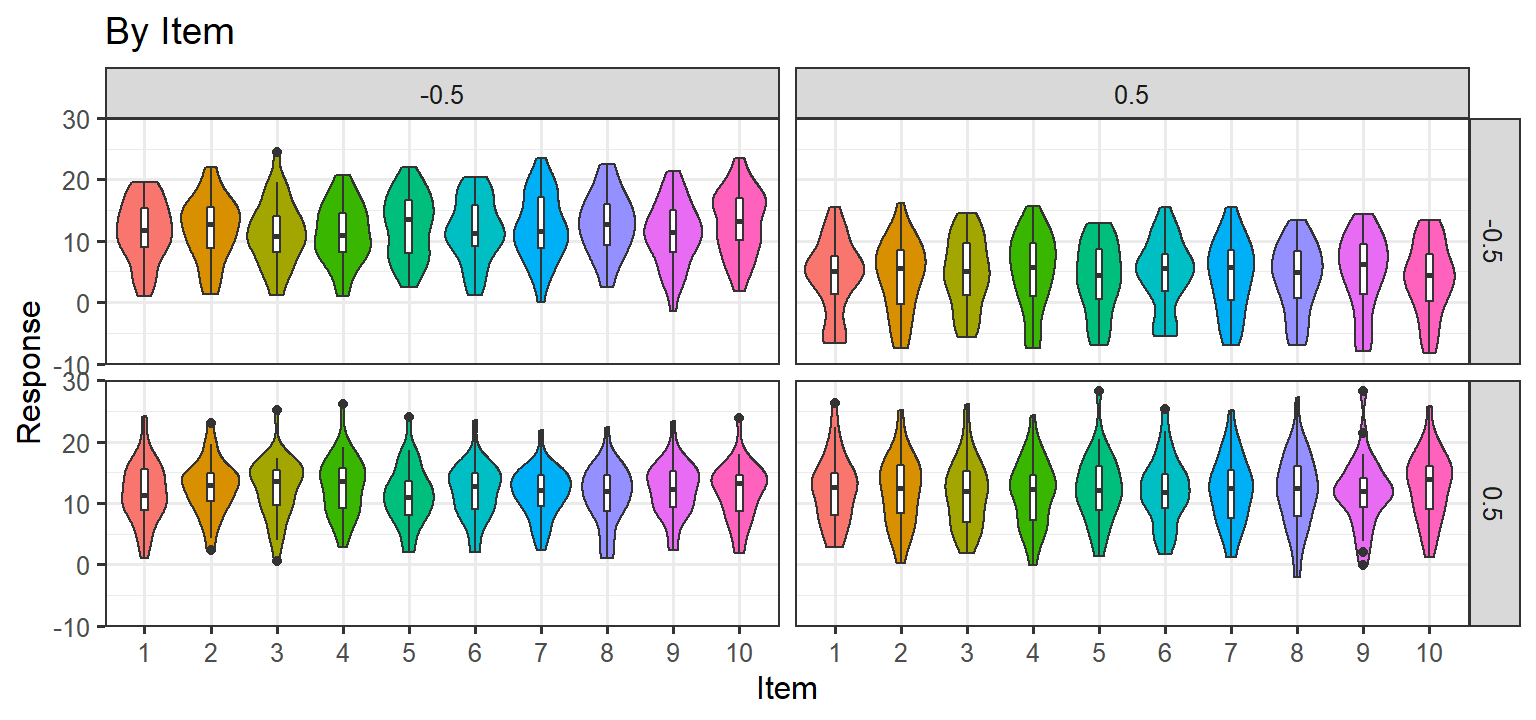
Parsimonious Stages
- Step 1:
(1+C1*C2|Subject) + (1+C1*C2|Item)
DV_SS_RSlope_SSNoise.M1<-lmer(DV_SS_RSlope_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1+Condition1*Condition2|Item),
data=DataSim8, REML=FALSE)
summary(DV_SS_RSlope_SSNoise.M1, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 | Subject) + (1 + Condition1 * Condition2 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4539.2 4666.4 -2244.6 4489.2 1175
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.8240 -0.6370 0.0118 0.6150 3.7034
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 23.436815 4.84116
## Condition1 4.416334 2.10151 -0.20
## Condition2 4.036480 2.00910 0.39 0.08
## Condition1:Condition2 13.515658 3.67636 0.03 0.27 0.05
## Item (Intercept) 0.006239 0.07898
## Condition1 0.022577 0.15026 0.15
## Condition2 0.029861 0.17280 -0.93 0.23
## Condition1:Condition2 2.707920 1.64558 0.39 -0.85 -0.71
## Residual 1.624477 1.27455
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0711 0.8850 30.0546 11.380 2.03e-12 ***
## Condition1 3.8278 0.3936 30.7373 9.726 6.77e-11 ***
## Condition2 -3.7412 0.3781 31.0274 -9.895 4.08e-11 ***
## Condition1:Condition2 7.5843 0.8620 33.4311 8.799 3.22e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- The model fit and everything seems to be OK, but random correlations on the items are bit high.
- Step 2: extract the PCA of the random structure from the maximal model
DV_SS_RSlope_SSNoise.M1.PCA<-rePCA(DV_SS_RSlope_SSNoise.M1)
summary(DV_SS_RSlope_SSNoise.M1.PCA)## $Subject
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 3.8711 2.9343 1.61133 1.32623
## Proportion of Variance 0.5361 0.3080 0.09289 0.06293
## Cumulative Proportion 0.5361 0.8442 0.93707 1.00000
##
## $Item
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 1.2988 0.12740 0.0001189 3.64e-20
## Proportion of Variance 0.9905 0.00953 0.0000000 0.00e+00
## Cumulative Proportion 0.9905 1.00000 1.0000000 1.00e+00For
Subject: we see all components capture some random variance, but the last term is rather a small amount. Maybe we don’t need the interaction (we will check this later)?For
Item: we see the first two components capture 100% of the random variance. So we really don’t need all that complexity.Step 3: Reduce complexity Cycle
Cycle 1
- Keep Subjects the same for now, but remove the random slopes per subject as 99% of the variance was captured by the random intercepts
DV_SS_RSlope_SSNoise.M2<-lmer(DV_SS_RSlope_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1+Condition1*Condition2||Item),
data=DataSim8, REML=FALSE)
summary(DV_SS_RSlope_SSNoise.M2, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 | Subject) + (1 + Condition1 * Condition2 || Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4534.5 4631.2 -2248.2 4496.5 1181
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.7800 -0.6295 0.0127 0.6215 3.5679
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 23.432281 4.84069
## Condition1 4.410880 2.10021 -0.20
## Condition2 4.034431 2.00859 0.39 0.08
## Condition1:Condition2 13.508926 3.67545 0.03 0.27 0.05
## Item (Intercept) 0.001747 0.04179
## Item.1 Condition1 0.000000 0.00000
## Item.2 Condition2 0.014630 0.12095
## Item.3 Condition1:Condition2 2.708513 1.64576
## Residual 1.639378 1.28038
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0711 0.8847 30.0274 11.384 2.03e-12 ***
## Condition1 3.8278 0.3905 30.0135 9.802 7.24e-11 ***
## Condition2 -3.7412 0.3760 30.3695 -9.949 4.52e-11 ***
## Condition1:Condition2 7.5843 0.8620 33.3288 8.799 3.31e-10 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- Recheck the PCA
DV_SS_RSlope_SSNoise.M2.PCA<-rePCA(DV_SS_RSlope_SSNoise.M2)
summary(DV_SS_RSlope_SSNoise.M2.PCA)## $Subject
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 3.8530 2.9208 1.60211 1.31972
## Proportion of Variance 0.5362 0.3081 0.09271 0.06291
## Cumulative Proportion 0.5362 0.8444 0.93709 1.00000
##
## $Item
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 1.285 0.09447 0.03264 0
## Proportion of Variance 0.994 0.00537 0.00064 0
## Cumulative Proportion 0.994 0.99936 1.00000 1- Still overparameterized for random effect on items
Cycle 2
- Remove condtion 2 from items
DV_SS_RSlope_SSNoise.M3<-lmer(DV_SS_RSlope_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1+Condition1||Item),
data=DataSim8, REML=FALSE)
summary(DV_SS_RSlope_SSNoise.M3, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 | Subject) + (1 + Condition1 || Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4617.4 4703.9 -2291.7 4583.4 1183
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9268 -0.6401 0.0307 0.6732 2.9642
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 2.343e+01 4.840e+00
## Condition1 4.394e+00 2.096e+00 -0.20
## Condition2 4.015e+00 2.004e+00 0.39 0.08
## Condition1:Condition2 1.326e+01 3.642e+00 0.03 0.27 0.05
## Item (Intercept) 2.245e-04 1.498e-02
## Item.1 Condition1 3.258e-09 5.708e-05
## Residual 1.822e+00 1.350e+00
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0711 0.8846 30.0105 11.38 2.05e-12 ***
## Condition1 3.8278 0.3906 29.9989 9.80 7.30e-11 ***
## Condition2 -3.7412 0.3741 29.9974 -10.00 4.56e-11 ***
## Condition1:Condition2 7.5843 0.6829 30.0137 11.11 3.75e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')- Recheck the PCA
DV_SS_RSlope_SSNoise.M3.PCA<-rePCA(DV_SS_RSlope_SSNoise.M3)
summary(DV_SS_RSlope_SSNoise.M3.PCA)## $Subject
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 3.6544 2.7471 1.51483 1.24734
## Proportion of Variance 0.5395 0.3049 0.09271 0.06286
## Cumulative Proportion 0.5395 0.8444 0.93714 1.00000
##
## $Item
## Importance of components:
## [,1] [,2]
## Standard deviation 0.0111 4.229e-05
## Proportion of Variance 1.0000 1.000e-05
## Cumulative Proportion 1.0000 1.000e+00- Model is still overparameterized for items
Cycle 3
- It seems we should only have intercepts only for items
DV_SS_RSlope_SSNoise.M4<-lmer(DV_SS_RSlope_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1|Item),
data=DataSim8, REML=FALSE)
summary(DV_SS_RSlope_SSNoise.M4, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 | Subject) + (1 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4615.4 4696.8 -2291.7 4583.4 1184
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9268 -0.6401 0.0307 0.6732 2.9642
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 2.343e+01 4.84031
## Condition1 4.393e+00 2.09593 -0.20
## Condition2 4.016e+00 2.00398 0.39 0.08
## Condition1:Condition2 1.326e+01 3.64178 0.03 0.27 0.05
## Item (Intercept) 2.215e-04 0.01488
## Residual 1.822e+00 1.34992
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0711 0.8846 30.0137 11.385 2.04e-12 ***
## Condition1 3.8278 0.3905 30.0100 9.802 7.25e-11 ***
## Condition2 -3.7412 0.3741 29.9897 -10.001 4.58e-11 ***
## Condition1:Condition2 7.5843 0.6829 30.0172 11.106 3.74e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## Model failed to converge with max|grad| = 0.00236296 (tol = 0.002, component 1)- Check fit
## Data: DataSim8
## Models:
## DV_SS_RSlope_SSNoise.M4: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 | Subject) + (1 | Item)
## DV_SS_RSlope_SSNoise.M3: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 | Subject) + (1 + Condition1 || Item)
## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
## DV_SS_RSlope_SSNoise.M4 16 4615.4 4696.8 -2291.7 4583.4
## DV_SS_RSlope_SSNoise.M3 17 4617.4 4703.9 -2291.7 4583.4 0 1 0.9982- Removing items did not hurt us
Cycle 4
- It seems we should remove the random interaction at subjects?
DV_SS_RSlope_SSNoise.M5<-lmer(DV_SS_RSlope_SSNoise ~ Condition1*Condition2
+(1+Condition1+Condition2|Subject)
+(1|Item),
data=DataSim8, REML=FALSE)
summary(DV_SS_RSlope_SSNoise.M5, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 +
## Condition2 | Subject) + (1 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4965.2 5026.2 -2470.6 4941.2 1188
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.86669 -0.66924 0.02599 0.64881 2.87445
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 23.411 4.838
## Condition1 4.304 2.075 -0.20
## Condition2 3.925 1.981 0.39 0.09
## Item (Intercept) 0.000 0.000
## Residual 2.719 1.649
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0711 0.8847 30.0032 11.384 2.05e-12 ***
## Condition1 3.8278 0.3905 30.0073 9.801 7.27e-11 ***
## Condition2 -3.7412 0.3740 30.0015 -10.002 4.55e-11 ***
## Condition1:Condition2 7.5843 0.1904 1109.9980 39.833 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## boundary (singular) fit: see help('isSingular')Items is gone now as well, but removing the interaction hurt us?
Check fit
## Data: DataSim8
## Models:
## DV_SS_RSlope_SSNoise.M5: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 + Condition2 | Subject) + (1 | Item)
## DV_SS_RSlope_SSNoise.M4: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 | Subject) + (1 | Item)
## npar AIC BIC logLik deviance Chisq Df
## DV_SS_RSlope_SSNoise.M5 12 4965.2 5026.2 -2470.6 4941.2
## DV_SS_RSlope_SSNoise.M4 16 4615.4 4696.8 -2291.7 4583.4 357.76 4
## Pr(>Chisq)
## DV_SS_RSlope_SSNoise.M5
## DV_SS_RSlope_SSNoise.M4 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Model 4 was better, but let’s try one more simplification
DV_SS_RSlope_SSNoise.M6<-lmer(DV_SS_RSlope_SSNoise ~ Condition1*Condition2
+(1+Condition1*Condition2||Subject),
data=DataSim8, REML=FALSE)
summary(DV_SS_RSlope_SSNoise.M6, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 || Subject)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4610.5 4656.3 -2296.2 4592.5 1191
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.92704 -0.62995 0.03026 0.67443 2.97098
##
## Random effects:
## Groups Name Variance Std.Dev.
## Subject (Intercept) 23.435 4.841
## Subject.1 Condition1 4.394 2.096
## Subject.2 Condition2 4.015 2.004
## Subject.3 Condition1:Condition2 13.267 3.642
## Residual 1.822 1.350
## Number of obs: 1200, groups: Subject, 30
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.0711 0.8847 30.0002 11.384 2.06e-12 ***
## Condition1 3.8278 0.3906 30.0000 9.801 7.30e-11 ***
## Condition2 -3.7412 0.3740 30.0001 -10.002 4.55e-11 ***
## Condition1:Condition2 7.5843 0.6830 30.0003 11.104 3.78e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Check fit
## Data: DataSim8
## Models:
## DV_SS_RSlope_SSNoise.M6: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 || Subject)
## DV_SS_RSlope_SSNoise.M4: DV_SS_RSlope_SSNoise ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 | Subject) + (1 | Item)
## npar AIC BIC logLik deviance Chisq Df
## DV_SS_RSlope_SSNoise.M6 9 4610.5 4656.3 -2296.2 4592.5
## DV_SS_RSlope_SSNoise.M4 16 4615.4 4696.8 -2291.7 4583.4 9.0836 7
## Pr(>Chisq)
## DV_SS_RSlope_SSNoise.M6
## DV_SS_RSlope_SSNoise.M4 0.2467- Model 6 is basically as good as model 4
Results of Simulation 2
- Model 6 seems best which makes sense given how we simulated the data, and we have cut the random parameters down by 7. You will notice the t-values from models 4 and 6 are very similar. Those parameters were not doing much for us. Also, items did not do anything for us, but that is logical given they did not vary.
Simulation 3: Random Slopes per subject + Noise/Item level Random Effects
- Here we let each subject have their own slope for each condition (1
& 2) and the interaction. We also add noise to each trial, and each
item has a random effect. Thus there are ARE random slopes per subject
and per item. and there are no item level effects. If this approach
matches the data, we will be left with this random structure:
(1+C1*C2|Subject) + (1+C1*C2|Item)
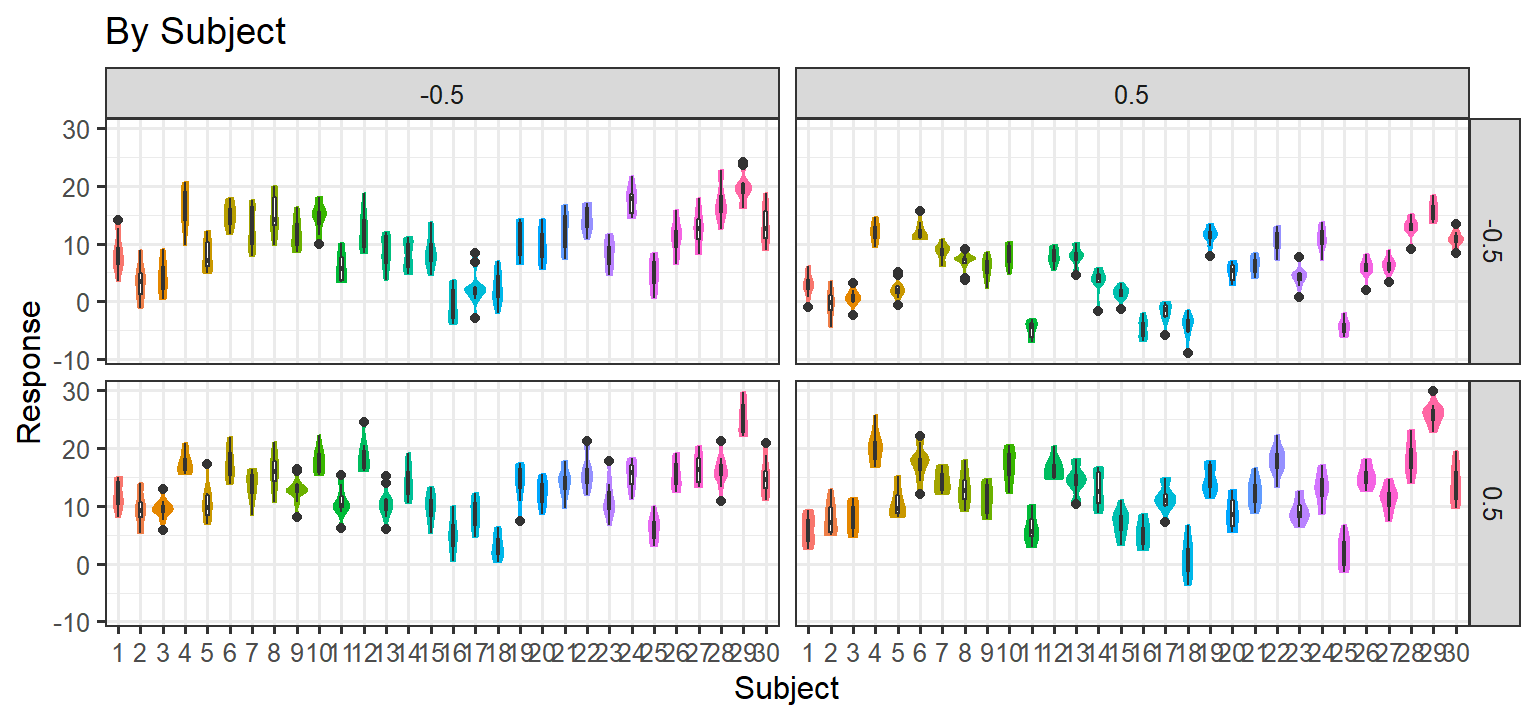
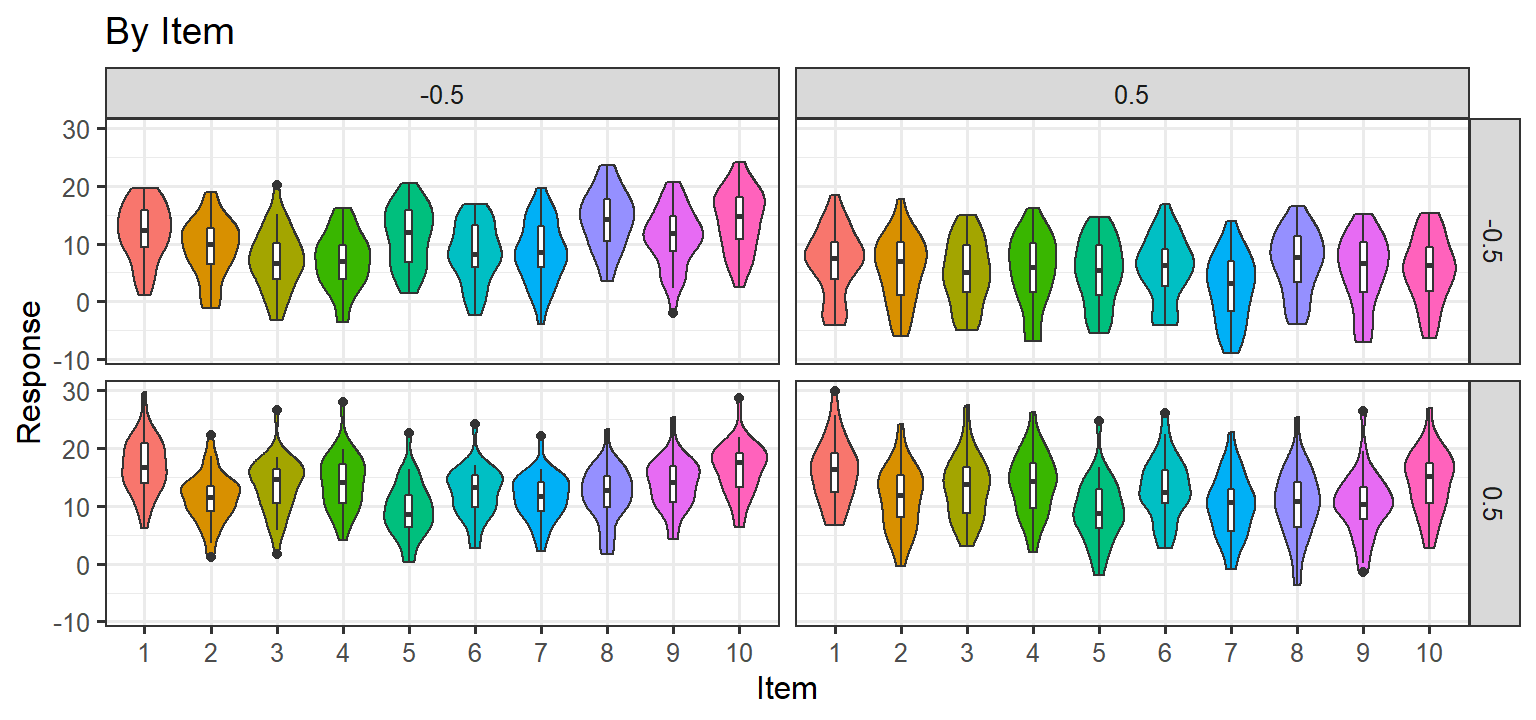
Parsimonious Stages
- Step 1:
(1+C1*C2|Subject) + (1+C1*C2|Item)
Max.M1<-lmer(DV_SS_RSlope_SSNoise_Items ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1+Condition1*Condition2|Item),
data=DataSim8, REML=FALSE)
summary(Max.M1, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula:
## DV_SS_RSlope_SSNoise_Items ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 | Subject) + (1 + Condition1 * Condition2 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4619.4 4746.6 -2284.7 4569.4 1175
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9978 -0.6342 0.0002 0.6443 3.5752
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 23.551 4.853
## Condition1 4.507 2.123 -0.20
## Condition2 4.096 2.024 0.38 0.09
## Condition1:Condition2 3.489 1.868 0.10 0.21 0.09
## Item (Intercept) 2.194 1.481
## Condition1 5.177 2.275 0.25
## Condition2 2.256 1.502 -0.32 0.47
## Condition1:Condition2 2.599 1.612 0.23 -0.60 -0.69
## Residual 1.640 1.281
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.1912 1.0029 39.2576 10.162 1.50e-12 ***
## Condition1 4.7773 0.8206 16.0111 5.822 2.59e-05 ***
## Condition2 -2.9977 0.6063 22.2226 -4.944 5.87e-05 ***
## Condition1:Condition2 3.9174 0.6309 18.3156 6.209 6.81e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
## optimizer (nloptwrap) convergence code: 0 (OK)
## Model failed to converge with max|grad| = 0.00415805 (tol = 0.002, component 1)- The model fit and everything seems to be OK, but random correlations on the items are bit high.
- Step 2: extract the PCA of the random structure from the maximal model
Max.M1.PCA<-rePCA(Max.M1)
summary(Max.M1.PCA)## $Subject
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 3.8626 1.7788 1.40911 1.29085
## Proportion of Variance 0.6864 0.1456 0.09135 0.07666
## Cumulative Proportion 0.6864 0.8320 0.92334 1.00000
##
## $Item
## Importance of components:
## [,1] [,2] [,3] [,4]
## Standard deviation 2.1269 1.4167 0.7041 0.65489
## Proportion of Variance 0.6068 0.2692 0.0665 0.05752
## Cumulative Proportion 0.6068 0.8760 0.9425 1.00000For
Subject&Item: we see all components capture some random variance.Step 3: Reduce complexity Cycle
Cycle 1
- The only thing I can try to simply is to remove the random correlations and see if it does not change the fit.
- Remove effects on items:
Max.M2<-lmer(DV_SS_RSlope_SSNoise_Items ~ Condition1*Condition2
+(1+Condition1*Condition2|Subject)
+(1+Condition1*Condition2||Item),
data=DataSim8, REML=FALSE)
summary(Max.M2, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula:
## DV_SS_RSlope_SSNoise_Items ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 | Subject) + (1 + Condition1 * Condition2 || Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4621.1 4717.8 -2291.5 4583.1 1181
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9004 -0.6446 0.0127 0.6317 3.5407
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 23.608 4.859
## Condition1 4.527 2.128 -0.20
## Condition2 4.115 2.028 0.38 0.08
## Condition1:Condition2 3.514 1.875 0.10 0.22 0.09
## Item (Intercept) 2.240 1.497
## Item.1 Condition1 5.197 2.280
## Item.2 Condition2 2.273 1.508
## Item.3 Condition1:Condition2 2.619 1.618
## Residual 1.640 1.281
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.1912 1.0061 38.8892 10.129 1.84e-12 ***
## Condition1 4.7773 0.8223 15.9698 5.810 2.68e-05 ***
## Condition2 -2.9977 0.6082 22.0265 -4.929 6.24e-05 ***
## Condition1:Condition2 3.9174 0.6332 18.1802 6.187 7.37e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Check fit
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq) Max.M2 19 4621.094 4717.806 -2291.547 4583.094 NA NA NA Max.M1 25 4619.369 4746.621 -2284.685 4569.369 13.72516 6 0.0328616 - Model 1 was better
- Next, remove random effects on Subjects:
Max.M3<-lmer(DV_SS_RSlope_SSNoise_Items ~ Condition1*Condition2
+(1+Condition1*Condition2||Subject)
+(1+Condition1*Condition2|Item),
data=DataSim8, REML=FALSE)
summary(Max.M3, corr=FALSE)## Linear mixed model fit by maximum likelihood . t-tests use Satterthwaite's
## method [lmerModLmerTest]
## Formula:
## DV_SS_RSlope_SSNoise_Items ~ Condition1 * Condition2 + (1 + Condition1 *
## Condition2 || Subject) + (1 + Condition1 * Condition2 | Item)
## Data: DataSim8
##
## AIC BIC logLik deviance df.resid
## 4615.5 4712.2 -2288.7 4577.5 1181
##
## Scaled residuals:
## Min 1Q Median 3Q Max
## -2.9901 -0.6311 0.0072 0.6479 3.5794
##
## Random effects:
## Groups Name Variance Std.Dev. Corr
## Subject (Intercept) 23.583 4.856
## Subject.1 Condition1 4.522 2.126
## Subject.2 Condition2 4.105 2.026
## Subject.3 Condition1:Condition2 3.494 1.869
## Item (Intercept) 2.212 1.487
## Condition1 5.194 2.279 0.26
## Condition2 2.267 1.506 -0.33 0.46
## Condition1:Condition2 2.606 1.614 0.23 -0.61 -0.69
## Residual 1.640 1.281
## Number of obs: 1200, groups: Subject, 30; Item, 10
##
## Fixed effects:
## Estimate Std. Error df t value Pr(>|t|)
## (Intercept) 10.1912 1.0043 39.1091 10.148 1.63e-12 ***
## Condition1 4.7773 0.8219 15.9698 5.812 2.67e-05 ***
## Condition2 -2.9977 0.6074 22.0934 -4.935 6.10e-05 ***
## Condition1:Condition2 3.9174 0.6316 18.2527 6.202 7.02e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1- Check fit
## Data: DataSim8
## Models:
## Max.M3: DV_SS_RSlope_SSNoise_Items ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 || Subject) + (1 + Condition1 * Condition2 | Item)
## Max.M1: DV_SS_RSlope_SSNoise_Items ~ Condition1 * Condition2 + (1 + Condition1 * Condition2 | Subject) + (1 + Condition1 * Condition2 | Item)
## npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
## Max.M3 19 4615.5 4712.2 -2288.7 4577.5
## Max.M1 25 4619.4 4746.6 -2284.7 4569.4 8.088 6 0.2317Model 3 is no worse a fit than Model 1.
Results of Simulation 3
- Model 3 is basically as good as model 1 (the maximal model), but we reduced the complexity and removed 6 random parameters. You will notice the t-values from models 1 and 3 are very similar. Those parameters were not doing much for us.